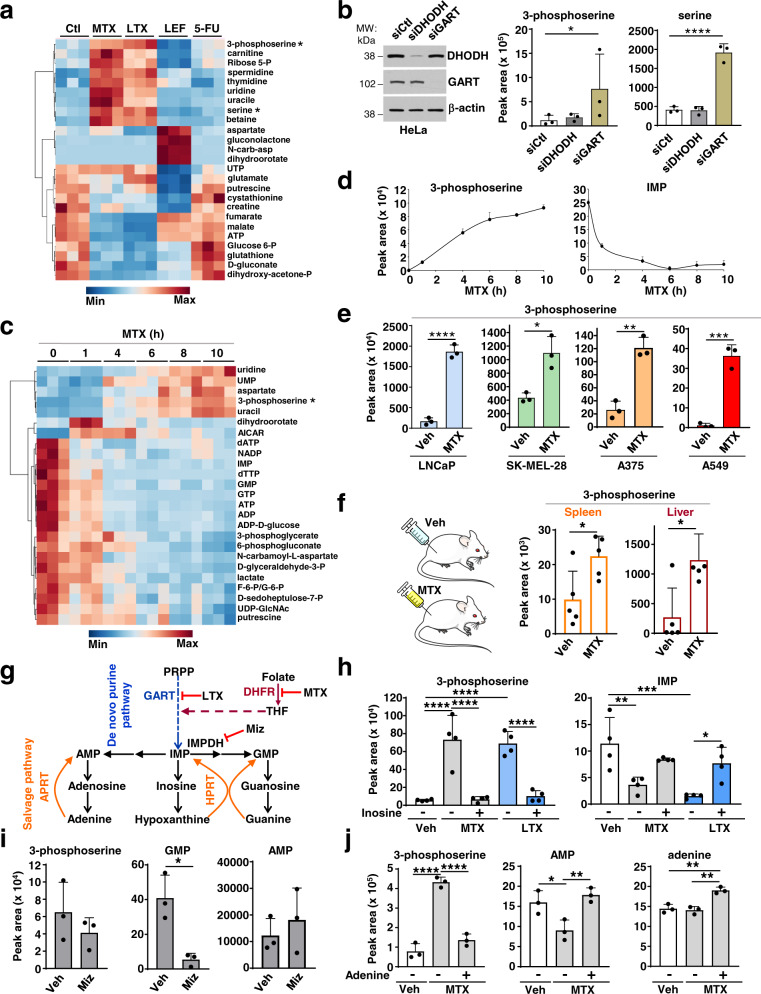
Fig. 2. Depletion of purines results in elevated 3-phosphoserine and serine levels.
a Steady-state metabolite profile from HeLa cells treated for 15 h with DMSO (Ctl), MTX (2 µM), LTX (2 µM), Lef (10 µM), and 5-FU (1 µM). b Normalized peak areas of 3-phosphoserine and serine measured by LC-MS/MS from HeLa cells transfected with nontargeting controls (siCtl), or siRNA against DHODH or GART. c As in (a) from HeLa cells treated with MTX (2 µM) for the indicated times. d Normalized peak areas of 3-phosphoserine and IMP from HeLa cells treated with MTX (2 µM) for the indicated times are shown from the experiment in (c). e Normalized peak areas of 3-phosphoserine after vehicle (Veh, DMSO) or MTX treatment (2 µM, 15 h) from LNCaP (prostate cancer), SK-MEL-28 (melanoma), A375 (melanoma), and A549 (lung cancer) cells. f Normalized peak areas of 3-phosphoserine from murine spleen and liver after daily treatment with vehicle (0.9% saline) or MTX (40 mg/kg) for five consecutive days. g Schematic of the de novo and salvage purine synthesis pathways. The targets of the purine synthesis inhibitors (MTX or LTX) and Mizoribine (Miz) are indicated. h Normalized peak areas of 3-phosphoserine and IMP after treatment of HeLa cells treated with vehicle (Veh, DMSO), MTX (2 µM), or LTX (2 µM) for 15 h, followed by treatment with inosine (50 µM, 1 h). i Normalized peak areas of the indicated metabolites from HeLa cells treated with vehicle (Veh, DMSO) or Mizoribine (Miz, 15 µM) for 15 h. j Normalized peak areas of the indicated metabolites from HeLa cells treated with vehicle (Veh, DMSO) or MTX (2 µM) for 15 h, followed by treatment with adenine (50 µM, 1 h). b, d–f, h–j Data are presented as the mean ± s.d from n = 3 (b, d, e, i, j) or n = 4 (h), or n = 5 (f) of biologically independent samples. Data are presented from n = 3 independent samples (a, c). b, d, h, j One-way ANOVA, Turkey’s post-hoc test, multiple comparison, e, f, i Unpaired t-test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are representative of n = 2 (a–d, f, i, j), n = 3 (e), n = 4 (h) independent experiments. Source data and exact p-values are provided as a Source Data file.