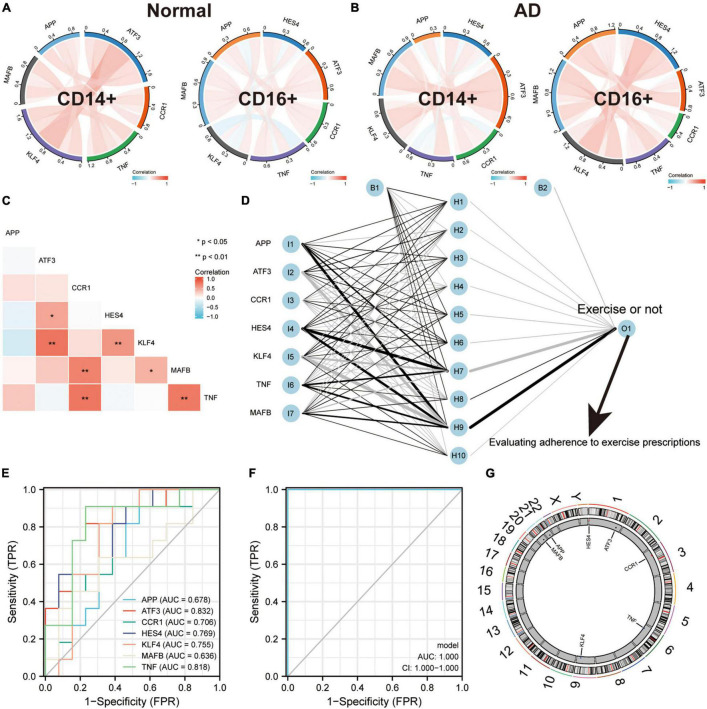
FIGURE 8.
ERADMT gene set as an indicator for assessing adherence to exercise prescription in patients with AD. (A,B) Correlation of APP, CCR1, TNF, ATF3, KLF4, HES4, and MAFB expression in CD14+ and CD16+ monocytes based on the GSE181279 single cell dataset, analyzed using Person method, including NC group (A) and AD group (B), red indicates positive correlation whereas blue indicates negative correlation; (C) Expression correlations of APP, CCR1, TNF, ATF3, KLF4, HES4, and MAFB in monocytes were analyzed using the Person method based on the GSE181279 bulk RNA dataset; (D) Schematic diagram of a neural network model for determining whether to exercise using APP, CCR1, TNF, ATF3, KLF4, HES4, and MAFB; (E) The predictive ability of the ERADMT gene set (APP, CCR1, TNF, ATF3, KLF4, HES4, and MAFB) to separately predict whether to undergo exercise or not; (F) ROC curve indicating that the neural network model can accurately predict patients’ exercise compliance; (G) The position of the ERADMT gene set (APP, CCR1, TNF, ATF3, KLF4, HES4, and MAFB) on the chromosome is indicated by the circle plot.