Fig. 3.
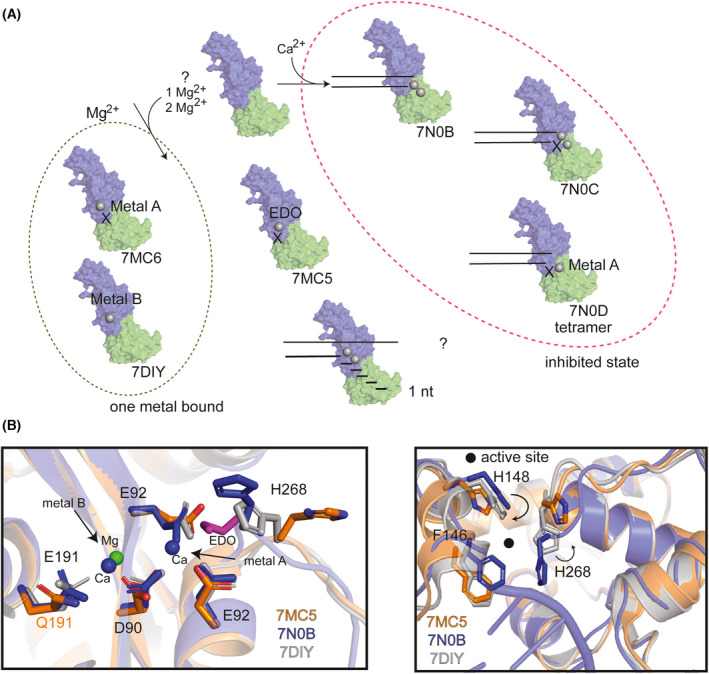
Nsp14 exonuclease cleavage mechanism. (A) Overview of current mechanistic analysis in structural studies with corresponding PDB IDs below the transparent cartoon surface representation of Nsp14. Dashed oval (green, left) structures only have one‐metal site bound with Mg2+. Dashed oval (magenta, right) indicates an inhibited (Ca2+‐bound states). Two parallel lines are a cartoon schematic of RNA. PDB ID: 7N0B was used to generate the surface presentation, which is colored by domain (ExoN, blue and MTase, green). Ethylene glycol (EDO). X indicates a mutated active site enzyme. (B) (left) Active site residues are shown in sticks and transparent cartoon presentation of the zoomed‐in exoN. Three PDBs were analyzed 7MC5 (orange), 7N0B (blue), and 7DIY (gray). 7MC5 has a E191Q mutation (orange text), all other residues written in black text and the EDO (purple) is shown in stick representation. Metal site locations are indicated with arrows. (right) Cartoon representation of PDBs globally aligned to 7DIY and movement is indicated by black arrows in the location of RNA binding. Select residues are highlighted by sticks and colored according to the PDB color. A filled black circle highlights the location of the active site.
