Fig. 5.
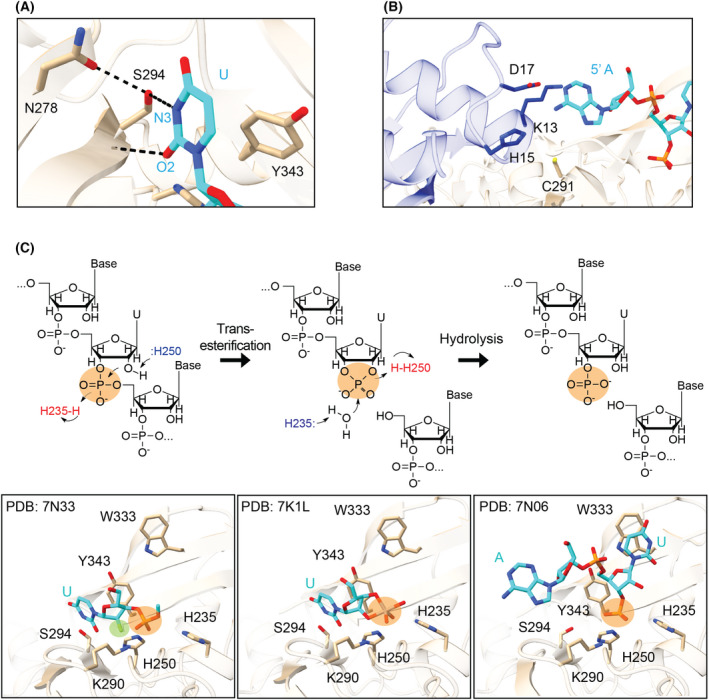
Nsp15 endoribonuclease cleavage mechanism. (A) Uridine discrimination occurs via a specific hydrogen bond network; N278 positions S294 to form a hydrogen bond with the N3 position of uridine. Y343 supports the position of the base through van der Waals interactions with the ribose sugar (PDB: 7N33). (B) A postcleavage structure showed interactions between the 5′ base and N‐terminal residues of a neighboring protomer (shown as blue sticks), mediated by water (PDB: 7N06). (C) The two‐step metal ion‐independent cleavage reaction shown in schematic form (top), with active site snapshots from structures of each state below (PDB: 7N33, 7K1L, 7N06) [49, 66]. Bottom: Nsp15 residues (tan) and RNA (cyan) are shown in stick format. The scissile phosphate is highlighted with an orange circle in both views. Prior to cleavage, the uridine is positioned for cleavage, interacting with the discriminatory residue S294. The position highlighted in green in the bottom left panel is the 2′ fluorine modification to prevent cleavage, showing the 2′ position is positioned in the catalytic triad for cleavage. Following the transesterification reaction, the uridine base and cyclic phosphate are positioned in the same places (central panel). After hydrolysis, however, the uridine base pivots to π‐stack with W333 (right panel).
