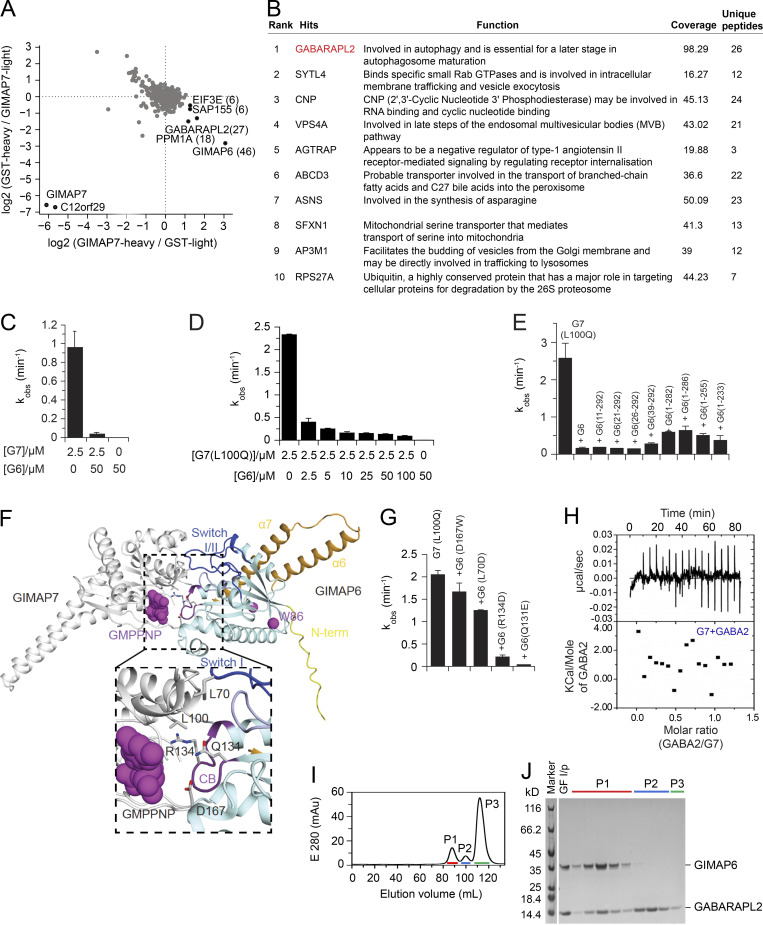
Figure S3.
GIMAP6, GABARAPL2, and GIMAP7 exist as functional complex. (A) SILAC based pull-down assays with GST-GIMAP7 (L100Q) or GST (control). The log2 fold changes of heavy to light ratio from forward and reverse experiments are plotted on the x and y axes, respectively. Proteins that had at least six identified peptides are plotted on the graph. The total number of peptides from specific interaction partners used for quantification in both forward and reverse experiments are indicated in brackets. Specific GIMAP7-interactors found in both experiments are located in the right lower corner. (B) Rank table of proteins with the highest overall coverage and unique peptides from immunoprecipitation of tandem-tagged GIMAP6 overexpressed in HEK293T cells followed by MS protein identification. The top ranked protein (GABARAPL2) is in red. (C) kobs for GTP hydrolysis of GIMAP6 (G6), GIMAP7 (G7), and the GIMAP6:GIMAP7 complex at the indicated concentrations at 20°C. (D) GTP hydrolysis of 2.5 μM GIMAP7 (L100Q) (G7 [L100Q]) in the presence of different GIMAP6 (G6) concentrations at 20°C. (E) GTP hydrolysis of 2.5 µM GIMAP7 (L100Q) upon addition of 50 µM of the N- or C-terminal deletion constructs of GIMAP6. In these experiments, the L100Q variant of GIMAP7 was used, since it has more stable aqueous solubility and can be purified at 10-fold higher yields than WT; in addition, it shows a two-fold increased GTPase activity compared to WT (Schwefel et al., 2013). (F) The AlphaFold 2 model of GIMAP6 was aligned on the GIMAP7 homodimer (PDB: 3ZJC) to obtain a model of the GIMAP6-GIMAP7 hetero-dimeric complex. GIMAP6 colors are as in Fig. 2 D. Switches I and II (blue), the P-loop (light blue), and the conserved box (CB, magenta) are involved in hetero-dimerization. Interface residues of GIMAP6 that were probed for GTPase interference of GIMAP7 in G and Leu100 in GIMAP7 are indicated. (G) GTP hydrolysis of 2.5 µM GIMAP7 (L100Q) upon addition of 50 µM of the indicated GIMAP6 mutants. (H) Isothermal titration calorimetry measurement indicate no binding of GABARAPL2 (GABA2) to GIMAP7 (G7). (I) Gel filtration run of the GIMAP6:GABARAPL2 complex on a Superdex200 column. (J) SDS-PAGE with selected fractions of the gelfiltration run in I. Peak 1 (P1) fractions were used for the subsequent GTP hydrolysis assays. GF I/P, protein applied to gel filtration. Bars (C–E and G) represent mean ± SEM. Data represent three experiments (C–E and G–J).