Abstract
Proper protein destruction by the ubiquitin (Ub)‐proteasome system is vital for a faithful cell cycle. Hence, the activity of Ub ligases is tightly controlled. The Anaphase‐Promoting Complex/Cyclosome (APC/C) is a 1.2 MDa Ub ligase responsible for mitotic progression and G1 maintenance. At the G1/S transition, the APC/C is inhibited by EMI1 to prevent APC/C‐dependent polyubiquitination of cell cycle effectors. EMI1 uses several interaction motifs to block the recruitment of APC/C substrates as well as the APC/C‐associated E2s, UBE2C, and UBE2S. Paradoxically, EMI1 is also an APC/C substrate during G1. Using a comprehensive set of enzyme assays, we determined the context‐dependent involvement of the EMI1 motifs in APC/C‐dependent ubiquitination of EMI1 and other substrates. Furthermore, we demonstrated that an isolated C‐terminal peptide fragment of EMI1 activates APC/C‐dependent substrate priming by UBE2C. Together, these findings reveal the multiple roles of the EMI1 C‐terminus for G1 maintenance and the G1/S transition.
Keywords: anaphase‐promoting complex/cyclosome, cell cycle, early mitotic inhibitor 1, ubiquitin conjugating enzyme E2 C, ubiquitin conjugating enzyme E2 S, ubiquitin ligase
1. INTRODUCTION
Ubiquitin (Ub)‐dependent protein turnover is paramount to the fidelity of the eukaryotic cell cycle. 1 Several Ub ligases target a vast array of protein substrates for polyubiquitination and subsequent degradation by the proteasome. Through this regulatory mechanism, checkpoint inhibitors are removed to promote cell cycle progression. 2 , 3 Disturbing or selectively modulating this system can result in the cause or treatment of cancer, respectively. 4 , 5
An essential, cell cycle‐regulated Ub ligase is the 1.2 MDa Anaphase‐Promoting Complex/Cyclosome (APC/C). 6 , 7 The APC/C has numerous notable cell cycle targets, such as securin and cyclin B, and is responsible for controlling the mitotic checkpoint and the maintenance of G1. 8 , 9 , 10 , 11 , 12 , 13 , 14 The APC/C is then shut down to permit S phase entry. 15 , 16 , 17 In addition to its roles in the cell cycle, the APC/C has explicit functions in development, differentiation, and metabolism in specific cell types, for example, neurons. 18 , 19 Due to its importance in this wide range of processes, APC/C activity is tightly controlled.
At the G1/S boundary, APC/C activity is halted by the phosphorylation and ubiquitination of its coactivator (CDH1) and also through the binding of the protein inhibitor EMI1 (reviewed in 20 ). At this critical checkpoint, the cell cycle may enter S phase. 17 The C‐terminus of EMI1 (~16 kDa) blocks APC/C function through multisite binding with various motifs. 21 , 22 , 23 , 24 , 25 Recent studies have shown that EMI1 alternates between being a substrate and inhibitor of the APC/C through multivalent interactions. 26 However, it is unclear how this process occurs mechanistically.
Similar to other RING E3 Ub ligases, the largest family of Ub ligases, the APC/C serves as a scaffold to simultaneously bind both a substrate and Ub‐conjugating enzymes (E2s). 7 , 27 , 28 APC/C recognizes substrates through short motifs called degrons, such as the destruction box (D‐box) and KEN‐box. 29 , 30 , 31 , 32 APC/C also recruits its associated E2s, UBE2C, and UBE2S, to facilitate Ub transfer: once bound to the APC/C catalytic module, UBE2C first primes the substrate with Ub on substrate lysines or forms short Ub chains, while UBE2S extends K11‐linked Ub chains. 8 , 10 , 13 , 33 , 34 , 35 , 36 UBE2C simultaneously binds and is activated by the cullin subunit APC2 winged‐helix B (WHB) and the APC11 RING domain. 37 , 38 UBE2S uses a distinct mechanism for Ub chain‐elongation where its C‐terminal peptide (CTP) docks at a groove formed by subunits APC2 and APC4, and the E2 core domain binds to a surface on APC2, distinct from UBE2C. 21 , 34 , 36 , 37 , 39 Recent work by our group has shown that the UBE2SCTP binds and activates the APC/C to function with UBE2C for substrate priming. 40
EMI1 disrupts nearly every step in APC/C‐mediated ubiquitination by blocking substrate recruitment and both E2 binding sites to minimize the probability of EMI1 becoming an APC/C substrate (Figure 1a,b). 21 , 22 , 25 , 41 , 42 Specifically, EMI1 contains a D‐box that binds to CDH1 and APC10, blocking canonical substrates (cyclin B and securin). 21 , 22 , 25 , 41 , 42 EMI1 also contains a Linker and Zinc‐binding region (ZBR) that binds to the UBE2C‐binding site on APC11. 21 , 22 , 42 At its extreme C‐terminus, EMI1 contains a sequence that is incredibly similar to the C‐terminus of UBE2S, allowing EMI1 to utilize the UBE2SCTP binding site. 21 , 22 , 25 , 37 These EMI1 motifs synergize to create a tight‐binding inhibitor of the APC/C, fully preventing ubiquitination and allowing S‐phase entry.
FIGURE 1.
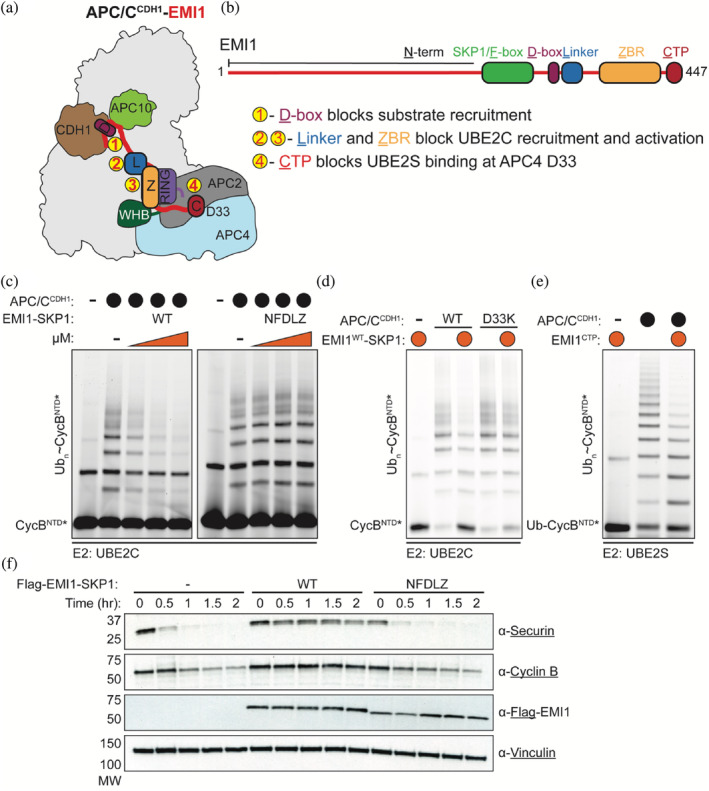
EMI1 C‐terminus is required for potent inhibition of APC/C‐dependent ubiquitination. (a) EMI1's D‐box, Linker, ZBR, and C‐terminal extension shut down multiple steps of APC/CCDH1‐dependent ubiquitination activity. Its D‐box docks between the APC/C coactivator CDH1 and APC10, preventing the binding of D‐box containing substrates. The Linker and ZBR interact with the APC11 RING domain, perturbing its interaction with the E2 UBE2C. Lastly, the C‐terminus binds to the APC2/APC4 groove, disrupting the recruitment of the E2 UBE2S. (b) Domain architecture of EMI1 and its multiple APC/C‐interacting motifs. (c) Elimination of the EMI1CTP (EMI1NFDLZ‐SKP1) disrupts the tight‐binding inhibition of EMI1WT‐SKP1 against APC/CCDH1‐UBE2C‐dependent reactions. Reactions were monitored by scanning of SDS‐PAGE gels where an N‐terminal fragment of Cyclin B (CycBNTD*) is fluorescently labeled. n = 3 independent experiments. (d) APC/C harboring a substitution in the APC2/APC4 groove (D33K) weakens the ability of EMI1WT‐SKP1 to inhibit APCCDH1‐UBE2C‐dependent reactions. Representative fluorescent scan of multiple (n > 3) independent experiments. (e) The addition of the EMI1CTP in isolation is sufficient to weaken APC/CCDH1‐UBE2S‐dependent polyubiquitination of a Ub‐fused fluorescent substrate (Ub‐CycBNTD*). n = 3 independent experiments. (f) EMI1WT‐SKP1 robustly prevents the destruction of APC/C substrates, followed by immunoblotting, in G1 extracts prepared from HeLa S3 cells. However, the EMI1NFDLZ‐SKP1 variant, a truncated version of EMI1 that lacks the EMI1CTP, restores the proteolysis of APC/C substrates. n = 3 independent experiments
The current model of how EMI1 transitions from an APC/C substrate to an inhibitor is through EMI1 accumulation, where EMI1 is a substrate at low concentrations but an inhibitor at high concentrations. 26 Using detailed enzyme, binding, and cell‐based assays, we interrogated the reciprocal regulation between APC/C and EMI1 and demonstrate how the different domains of EMI1 contribute to its ubiquitination by the APC/C. Competition by either substrate or E2 binding has differential effects on the levels of EMI1 ubiquitination. Furthermore, given the high sequence homology between the C‐termini of UBE2S and EMI1, we found that the EMI1CTP stimulated APC/C‐UBE2C‐dependent substrate priming. Taken together, our data suggest that the multivalency of EMI1 is vital to its regulation and is used to tune APC/C function in a context‐dependent manner. These findings have implications for how proper G1 maintenance and the G1/S transition are achieved.
2. RESULTS
2.1. Successful inhibition of APC/C‐mediated substrate ubiquitination by EMI1 requires its extreme C‐terminus
EMI1 requires multiple domains (D‐box, linker, ZBR, and the C‐terminus) to bind tightly to APC/CCDH1, and each domain is required to shut down specific steps of APC/C‐dependent polyubiquitination. 21 , 22 , 25 To assess how the C‐terminus of EMI1 is involved in APC/C inhibition, we reconstituted substrate polyubiquitination by APC/CCDH1 and its inhibition by EMI1 using a recombinant system. First, we evaluated the tight‐binding inhibition of EMI1WT‐SKP1 (SKP1 was used to stabilize versions of EMI1 that contained an F‐box) by monitoring APC/CCDH1‐UBE2C‐mediated ubiquitination of the N‐terminal domain of human Cyclin B (CycBNTD*, * denotes fluorescent label and position). EMI1WT‐SKP1 potently inhibited APC/CCDH1‐mediated substrate ubiquitination, validating our recombinant system (Figure 1c). Importantly, the EMI1 variant lacking the extreme C‐terminal residues (EMI1NFDLZ‐SKP1) was severely defective in shutting down polyubiquitination by APC/CCDH1 and UBE2C (Figure 1c). Similarly, a single mutation (APC4 D33K) in a groove formed by APC2/APC4, where the C‐termini of EMI1 and UBE2S bind the APC/C, reduced EMI1‐mediated inhibition (Figure 1d). The EMI1 extreme C‐terminus is not only important for the potent inhibition of APC/CCDH1‐UBE2C‐dependent processes, but a peptide fragment of the EMI1 C‐terminus (EMI1CTP, residues 430–447) also prevents Ub chain‐elongation by APC/CCDH1‐UBE2S (Figure 1e). Next, we used a HeLa cell extract system to monitor APC/CCDH1‐dependent substrate degradation as a readout for APC/CCDH1 activity. In the presence of EMI1WT‐SKP1, substrate degradation was completely blocked compared to the addition of EMI1NFDLZ‐SKP1, where APC/CCDH1‐dependent substrate degradation is only slightly reduced (Figure 1f). Taken together, these assays demonstrate that the C‐terminus of EMI1 is required for its potent inhibition of both UBE2C‐and UBE2S‐dependent substrate polyubiquitination processes. Since it is known that the C‐terminal peptide of UBE2S (UBE2SCTP) binds to the same site on the APC/C as the EMI1CTP and stimulates APC/CCDH1‐UBE2C function, we postulated that the EMI1CTP could also be involved in APC/CCDH1‐dependent ubiquitination of EMI1 and other substrates.
2.2. The EMI1 C‐terminus restricts APC/C‐dependent EMI1 ubiquitination
Initial studies of EMI1 suggested that EMI1 is a substrate only when the C‐terminal ZBR and peptide are deleted. 42 More recently, it was proposed that EMI1 is a substrate at low concentrations in early G1 phase and an inhibitor at higher concentrations at the G1/S boundary. 26 To examine the role of individual EMI1 domains required for the switch from a substrate to inhibitor, EMI1 full‐length and truncations were purified, fluorescently labeled, and tested as substrates in APC/CCDH1‐UBE2C‐dependent ubiquitination reactions (Figure 2a). We uncovered that *EMI1WT‐SKP1 and the 16 kDa C‐terminus of EMI1 (*EMI1DLZC) are poorly ubiquitinated in our conditions (Figure 2b). This finding is not particularly surprising as the C‐terminal fragment contains all the domains known to block APC/C substrate ubiquitination. However, the version of EMI1‐SKP1 lacking the C‐terminus (*EMI1NFDLZ‐SKP1) was more efficiently ubiquitinated (Figure 2b). This result suggests that the extreme C‐terminus (residues 433–447) binding to the APC2/APC4 groove limits EMI1 ubiquitination.
FIGURE 2.
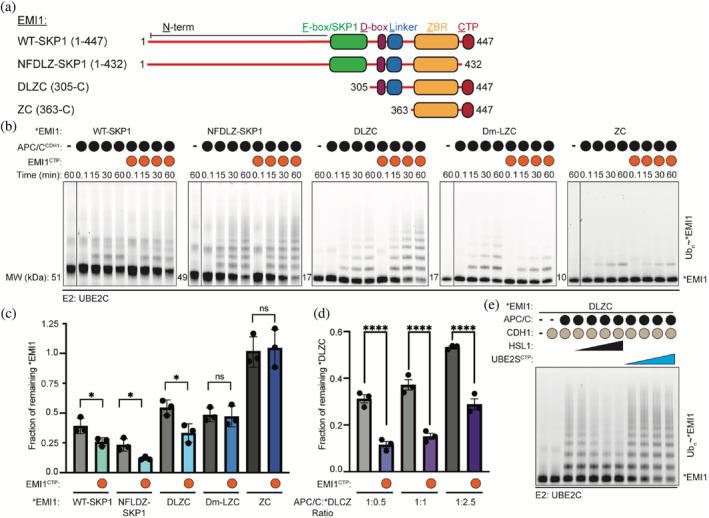
Multivalent binding of EMI1 requires its C‐terminal extension to prevent APC/CCDH1‐UBE2C‐dependent ubiquitination. (a) Schematic of the EMI1 truncations used in APC/CCDH1‐dependent ubiquitination assays. (b) APC/CCDH1‐UBE2C‐dependent ubiquitination of *EMI1WT‐SKP1, *EMI1NFDLZ‐SKP1, *EMI1DLZC, *EMI1Dm‐LZC, and *EMI1ZC was monitored by fluorescent scanning of SDS‐PAGE gels. The addition of the EMI1CTP in trans enhanced the ubiquitination of *EMI1WT‐SKP1, *EMI1NFDLZ‐SKP1, and *EMI1DLZC. n = 3 independent experiments. (c) Quantification of the amounts of *EMI1 remaining at the 60‐min time point. n = 3 independent experiments, ± SD (Unpaired t‐test, * p ≤ .05). (d) Similar reactions as in b, where the APC/CCDH1‐*EMI1DLZC ratios were varied. The amount of unmodified *EMI1DLZC at the 15‐minute time point was quantified. n = 3 independent experiments, ± SD (One‐way ANOVA, ****p ≤ .0001). (e) Titrating an APC/C substrate (HSL1 residues 768–842) prevents the APC/CCDH1‐UBE2C‐dependent ubiquitination of *EMI1DLZC, while the UBE2SCTP increases the depletion of unmodified EMI1DLZC. Reactions were monitored by fluorescent scanning and SDS‐PAGE, n = 3 independent experiments
To further test this hypothesis, the EMI1 truncations were tested in APCCDH1‐UBE2C‐dependent ubiquitination assays in the presence or absence of the isolated EMI1CTP. The addition of the EMI1CTP subtly enhanced the ubiquitination of *EMI1WT‐SKP1 and *EMI1NFDLZ‐SKP1 (Figure 2b,c). The increase in the rate of *EMI1 depletion and ubiquitination through the addition of the EMI1CTP was more noticeable in the *EMI1DLZC construct. This stimulatory effect was consistent at multiple APC/CCDH1‐EMI1 ratios (Figure 2d). Together, these results suggest that the EMI1CTP competes with the C‐terminal extension present on EMI1, promoting the depletion of unmodified EMI1. These results further indicate that the multivalent binding of EMI1WT to the APC/C facilitates the inhibition of both its own ubiquitination and ubiquitination of other APC/C substrates.
D‐boxes found in both EMI1 and dozens of APC/C substrates bind to a receptor site formed by CDH1 and APC10. 21 , 22 , 25 , 30 , 31 Because of the multivalent interactions between APC/CCDH1 and EMI1, we next examined the requirement of the EMI1 D‐box for EMI1 ubiquitination. Indeed, mutation of the EMI1 D‐box (*EMI1Dm‐LZC) or the removal of the EMI1 D‐box and linker (*EMI1ZC) dramatically reduced their ubiquitination compared to *EMI1DLZC (Figure 2b–d). Further, the titration of an APC/C substrate HSL1 (residues 768–842) from Saccharomyces cerevisiae, which contained both a D‐ and KEN‐box, reduced *EMI1DLZC ubiquitination, as well (Figure 2e). These results demonstrate that the EMI1 D‐box is important for its ubiquitination and suggests that the level of canonical substrates in cells may impact EMI1 ubiquitination.
UBE2S functions as the chain‐elongating E2 for the APC/C and its C‐terminus binds to the same APC2/APC4 groove as EMI1. 21 , 22 , 25 , 37 Therefore, we tested whether the UBE2SCTP, in isolation from its catalytic domain, promotes *EMI1DLZC ubiquitination. In support of this hypothesis, substrate depletion of *EMI1DLZC was stimulated by the addition of the UBE2SCTP (Figure 2e). Therefore, UBE2S has multiple functions during EMI1 ubiquitination, and its levels decline during G1 progression while the level of EMI1 increases. Taken together, our observations present an interesting hypothesis that UBE2S availability modulates the abundance of EMI1 as the UBE2SCTP and the EMI1CTP compete for the APC2/APC4 binding groove.
2.3. The EMI1CTP facilitates APC/C‐mediated substrate priming by UBE2C similar to the UBE2SCTP
The finding that the EMI1CTP promoted the ubiquitination of the EMI1 construct void of the EMI1 C‐terminus (EMI1NFDLZ‐SKP1) suggests that the EMI1CTP promotes APC/CCDH1‐UBE2C‐dependent ubiquitination. We previously described how the UBE2SCTP activates the APC/C through a mechanism distinct from CDH1‐induced activation. 40 Together, these findings led us to predict that the EMI1CTP, which shares 44% sequence identity with UBE2SCTP, could also facilitate UBE2C recruitment and activation when separated from the other EMI1 domains that inhibit APC/C function (Figure 3a). Remarkably, the EMI1CTP does behave in a manner analogous to the UBE2SCTP. First, the EMI1CTP accelerates the rate of UBE2C‐dependent Ub ligation to substrates and increases substrate depletion (Figure 3b,c). Second, to specifically characterize the first Ub transfer step (substrate priming), a single‐encounter assay was performed using methylated Ub (meUB) and a single lysine version of *CycBNTD(1K; Figure 3d). In this experiment, *CycBNTD(1K) was preincubated with APC/CCDH1 in the presence or absence of EMI1CTP or UBE2SCTP as a positive control. A mixture of UBE2C ~ Ub and excess unlabeled substrate (HSL1) were added. As a result, only modification of the prebound *CycBNTD(1K) would be observed with a single methyl‐Ub. Indeed, the EMI1CTP enhanced substrate priming by ~3‐fold, indicating that the EMI1CTP is activating the APC/C for UBE2C‐dependent priming (Figure 3e).
FIGURE 3.
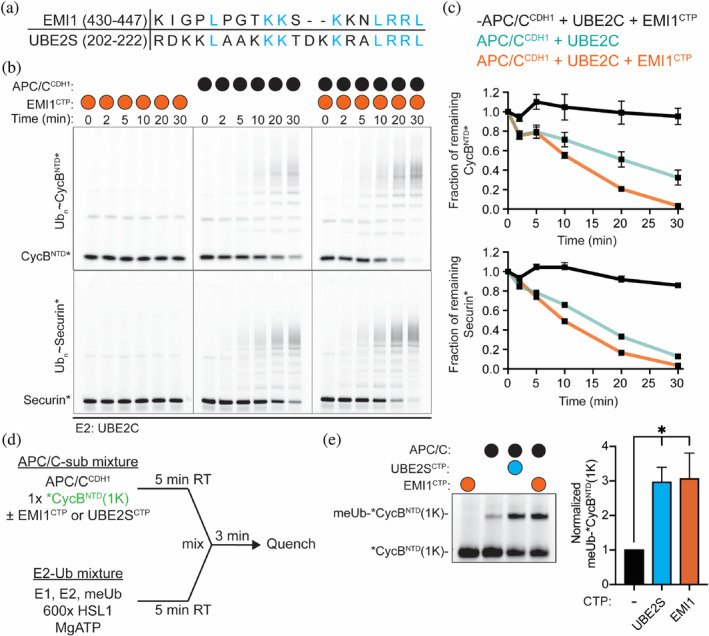
A peptide derived from the C‐terminus of EMI1 (EMI1CTP) enhances the rate of substrate modification by APC/CCDH1‐UBE2C. (a) Sequence alignment of the highly similar C‐termini of EMI1 and UBE2S. (b) Representative fluorescent image of SDS‐PAGE gels demonstrating that the addition of the EMI1CTP to APC/CCDH1‐dependent substrate polyubiquitination reactions (CycBNTD*, top, and Securin*, bottom) stimulates the rate of substrate depletion. (c) Quantification of the unmodified substrate from b. n = 3 independent experiments, ± SEM. (d) Schematic of the experimental set‐up of single‐encounter assays to monitor substrate priming by APC/CCDH1‐UBE2C used in e. In short, one mixture contains the components to load UBE2C with methylated Ub (E1, UBE2C, meUB, Mg/ATP) and an excess of unlabeled substrate. A second mixture contains the fluorescently labeled, single lysine version of CycB (*CycBNTD(1K)), APC/CCDH1, and either the EMI1CTP or the UBE2SCTP. Therefore, when the mixtures are combined, only the modification of the prebound substrate is monitored by fluorescent scans of SDS‐PAGE gels. (e) Representative SDS‐PAGE gel, left, demonstrating the addition of the EMI1CTP has a similar effect on APC/CCDH1‐dependent substrate priming as the UBE2SCTP. Right, quantification of the conjugated meUb‐*CycBNTD(1K). n = 3 independent experiments, ±SEM (Unpaired t‐test, *p ≤ .05)
2.4. The EMI1CTP activates the APC/C in a manner that is distinct from CDH1
An APC/C coactivator serves two different functions. First, it recruits substrates to the APC/C for ubiquitination. Second, it mobilizes the catalytic core (APC2–APC11), facilitating the recruitment and activation of UBE2C~Ub. This activation mechanism is dependent on a WD40 domain present in the APC1 subunit (Figure 4a). 43 To test if the EMI1CTP activates the APC/C similar to the UBE2SCTP rather than CDH1, we purified the recombinant APC/C harboring a deletion of this APC1 WD40 domain (APC/CΔAPC1‐WD40) and tested it against multiple substrates. As expected, the deletion of this domain significantly reduced the activity of APC/CCDH1‐dependent ubiquitination (Figure 4b,c, compare lanes 2 and 3). However, the UBE2SCTP enhances the ubiquitination of this otherwise defective APC/CΔAPC1‐WD40 variant (Figure 4b,c, compare lanes 3 and 4). By substituting the last 4 residues, which are conserved between both EMI1 and UBE2S (the LRRL residues), to alanine (UBE2SCTP‐4A), APC/C‐dependent function is greatly reduced (Figure 4b,c, compare lanes 4 and 5). However, the EMI1CTP rescues the enzymatic activity of the defective APC/CΔAPC1‐WD40 variant, which cannot be stimulated by CDH1, for all substrates tested including CycA*, CycBNTD*, Securin*, Ub‐CycBNTD*, and Ub‐Securin* (Figure 4b,c, compare lanes 3 and 6). Taken together, these data support the idea that the EMI1CTP assists in the recruitment of UBE2C to the APC/C for priming and multiubiquitination in a manner distinct from coactivators. This function is an interesting example of how an inhibitor captures and paradoxically induces the active state of a massive enzyme.
FIGURE 4.
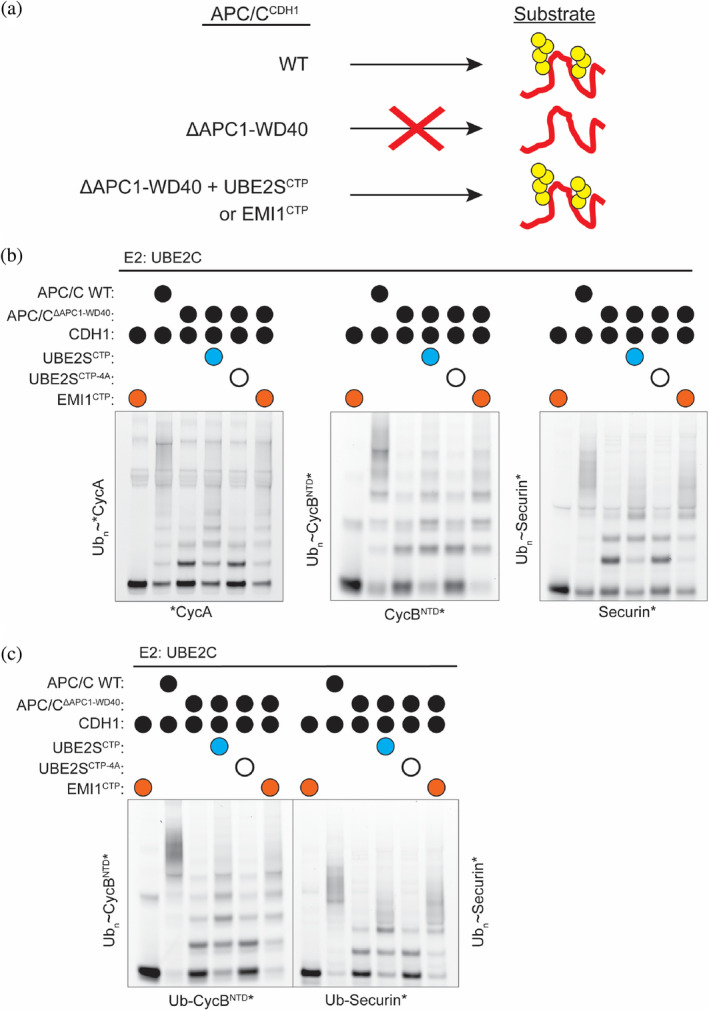
The EMI1CTP activates the APC/C through a different mechanism than the coactivator. (a) Removal of the WD40 domain of APC1 from the APC/C (APC/CΔAPC1‐WD40) prevents the mobilization of the catalytic core (APC2‐APC11) by the coactivator CDH1 for UBE2C function. However, the UBE2SCTP and the EMI1CTP activate this otherwise defective version of the APC/C. (b,c) APC/CCDH1‐UBE2C‐dependent polyubiquitination of substrates (b, CycA*, CycBNTD*, Securin*) and Ub‐modified substrates (c, Ub‐CycBNTD*, Ub‐Securin*), monitored by fluorescent scanning of SDS‐PAGE gels. This activity is markedly reduced when in the APC/CΔAPC1‐WD40 variant but is reestablished when either the UBE2SCTP or the EMI1CTP is added to the reaction. n = 3 independent experiments
3. DISCUSSION
The complex regulation of the APC/C requires the precise coordination of multiple cell cycle regulators. APC/C activity is high in late mitosis and early G1 but is reduced in late G1. 20 At the G1/S transition, EMI1 uses several domains to shut down nearly every aspect of ubiquitination by blocking the recruitment of the E2s, UBE2C, and UBE2S, and APC/C substrates. 21 , 22 , 25 Paradoxically, EMI1 is an APC/C substrate in early G1. 26 Therefore, how EMI1 changes from an APC/C substrate to an inhibitor has remained mechanistically unclear but important for a swift transition into S phase. This switch is significant as both the regulation of the G1/S transition and the protein levels of EMI1 are perturbed in many types of cancer. 44 , 45 , 46 , 47 , 48 , 49 As a whole, dysregulation of G1/S increases proliferation through a variety of mechanisms. 48 , 50 Similarly, EMI1 overexpression shortens G1, is a poor prognostic marker in solid tumors, and is considered oncogenic. 17 , 46 , 51 , 52 Here, we investigate the fundamental interactions between the APC/C and EMI1 to dissect how the APC/C recognizes EMI1 as a substrate and how different domains of EMI1 can modulate APC/C function.
The multivalency of EMI1 is foundational to its regulation by the APC/C. While previous studies have focused on how EMI1 uses its D‐box, Linker, ZBR, and C‐terminal peptide (CTP) for the tight‐binding inhibition of APC/C activity, here we flip the perspective and examine the individual contributions of these domains towards its ubiquitination. 21 , 22 , 25 , 26 , 41 , 42 For example, EMI1 ubiquitination is reduced by the addition of other substrates and by the engagement of the EMI1 C‐terminus to the groove formed by APC2/APC4. Unexpectedly, we revealed that the EMI1CTP in isolation can activate APC/CCDH1‐UBE2C‐dependent substrate priming. This activation seems surprising but may also function to accelerate substrate ubiquitination when UBE2S protein levels are low (Figure 5). Alternatively, the expected mobilization of the catalytic core may help the EMI1ZBR engage the APC11 RING domain, as the ZBR has little impact on substrate ubiquitination in isolation. 22
FIGURE 5.
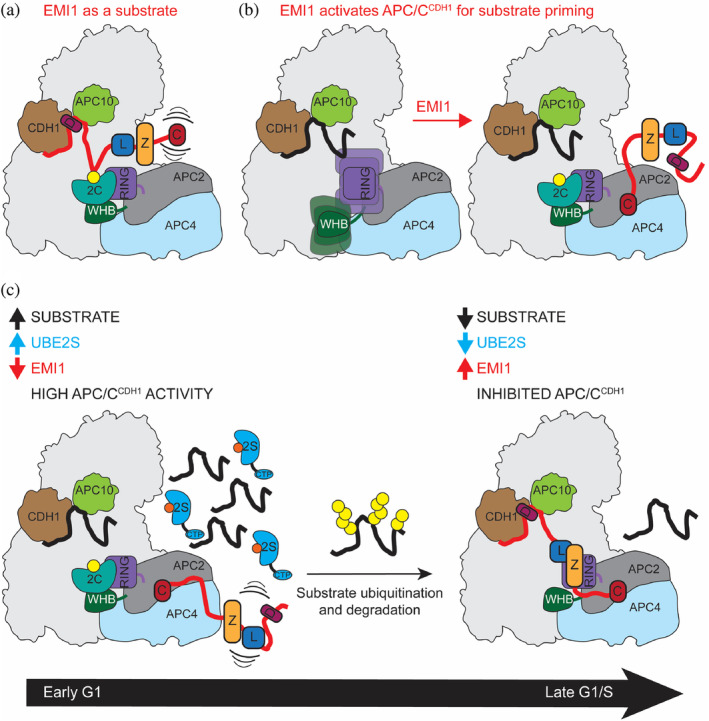
Multiple roles of the APC/C binding domains of EMI1 and its implications for context‐dependent modulation of APC/C activity. (a) EMI1 is recruited as a D‐box dependent substrate when its C‐terminus, which binds to the E2‐binding sites on the APC/C, is not engaged. (b) EMI1CTP binding to the APC2/APC4 groove stimulates APC/CCDH1‐UBE2C‐dependent substrate priming when the EMI1 D‐box is not bound to CDH1. (c) EMI1 binds to the APC/C via its EMI1CTP and may stimulate the ubiquitination of APC/C substrates in early G1 when the protein levels of other APC/C substrates and UBE2S are high. In late G1, when APC/C substrate levels are lower, the EMI1 D‐box can also engage APC/CCDH1 and fully inhibit the APC/C for the G1/S transition
Our data also demonstrate how levels of other APC/C substrates and UBE2S differentially impact EMI1 ubiquitination. The D‐box of EMI1 is important for its ubiquitination. Therefore, at high APC/C substrate levels in G1, EMI1 would compete for binding to the D‐box binding site, reducing EMI1 ubiquitination. As APC/C substrates are degraded throughout G1 progression, EMI1 is increasingly able to bind the D‐box binding site (Figure 5). UBE2S likely needs to be depleted during G1 to promote the role of EMI1 as an inhibitor rather than a substrate, because UBE2S has three roles during EMI1 ubiquitination: elongating K11‐linked Ub chains, activating the APC/C to function with UBE2C, and competing with EMI1 for binding to the APC2/APC4 groove to make EMI1 a better substrate. Taken together, our data reveal how the multivalency of EMI1 tunes its own ubiquitination and the activity of the APC/C.
4. MATERIALS AND METHODS
4.1. Protein purification
All proteins were expressed and purified as previously described. 22 , 40 In short, APC/C, CDH1, and UBA1 were expressed using a baculoviral expression system in High Five™ insect cells (Sigma‐Aldrich). EMI1 versions that contained an F‐box were co‐expressed and co‐purified with SKP1 for increased stability from insect cells. All other proteins for ubiquitination reactions were expressed in BL21‐Codon Plus (DE3)‐RIL cells. All purification steps were performed at 4°C and completed by size‐exclusion chromatography as the last step into a standard buffer, 20 mM HEPES pH 8.0, 200 mM NaCl, and 1 mM DTT, unless otherwise previously stated. All proteins were stored flash frozen in liquid nitrogen and stored at −80°C.
Fluorescein‐5‐maleimide was used to fluorescently label single cysteine versions of CycBNTD and Securin. CycBNTD(1K), Cyclin A, and EMI1 proteins were fluorescently labeled at their N‐terminus through a sortase‐mediated reaction. After TEV cleavage, these proteins were subjected to 10 nM–1 μM sortase and 400 μM–1 mM of a fluorescent‐labeled peptide (*LPETGG, * denotes fluorescent label) in 10 mM HEPES pH 8, 50 mM NaCl, 10 mM CaCl2 during an overnight incubation at 4°C. The fluorescently labeled proteins were purified by ion exchange and size‐exclusion chromatography.
4.2. Ubiquitination assays
Enzyme assays monitoring APC/C‐dependent ubiquitination were all performed at room temperature and quenched with SDS sample loading buffer. Reaction products were separated by SDS‐PAGE and visualized by fluorescent monitoring by an Amersham Typhoon Biomolecular imager. Quantitative assays were quantified using ImageQuant TL and statistically analyzed using GraphPad Prism v9.0.
Qualitative assays monitoring the ubiquitination of fluorescently labeled CycB (CycBNTD*) in the presence of a titration of EMI1 variants (WT and NFDLZ) were conducted by combining a final concentration of 1 μM UBA1, 5 mM MgCl2/ATP, 0.5 μM UBE2C, 80 nM APC/C, 0.5 μM CDH1, 0.5 μM CycBNTD*, a 3‐point titration of EMI1 WT or EMI1NFDLZ (80, 160, 310 nM), and 125 μM Ub. Reactions were quenched at 2.5 min (Figure 1c). Qualitative assays assessing the effect of APC4‐D33K mutation on EMI1‐dependent APC/C inhibition were performed by combining a final concentration of 1 μM UBA1, 5 mM MgCl2/ATP, 0.2 μM UBE2C, 0.2 μM CycBNTD*, 1 μM CDH1, 100 nM APC/C WT or D33K, 0.5 μM EMI1WT‐SKP1, and 125 μM Ub. Reactions were quenched at 10 min (Figure 1d). Qualitative assays probing the role of the isolated EMI1CTP on the inhibition of APC/CCDH1‐UBE2S dependent activity were performed by combining a final concentration of 1 μM UBA1, 5 mM MgCl2/ATP, 0.2 μM UBE2S, 200 nM APC/C WT, 0.5 μM CDH1, 0.2 μM substrate Ub‐CycBNTD*, 10 μM EMI1CTP, and 125 μM Ub. Reactions were quenched at 10 min with SDS buffer (Figure 1e). Assays monitoring the stimulation of APC/CCDH1‐UBE2C‐dependent substrate priming by the EMI1CTP (Figures 3 and 4) were performed as previously described, 40 except the UBE2SCTP was replaced by the EMI1CTP as indicated.
Assays monitoring the APC/C‐dependent ubiquitination of truncated EMI1 variants were conducted similar to the assays above. To determine the impact of the different domains of EMI1 and the EMI1CTP (KIGPLPGTKKSKKNLRRL), EMI1 ubiquitination assays were conducted by combining 0.5 μM UBA1, 0.5 μM UBE2C, 80 nM APC/C, 350 nM CDH1, 125 μM UB, 5 mM MgCl2/ATP, 200 nM of the respective fluorescent EMI1 variant, and either 25 μM EMI1CTP peptide or 20 mM HEPES pH 8, 200 mM NaCl buffer. Reactions were quenched at the indicated time points (Figure 2b–d). In Figure 2d, the *EMI1DLZC concentrations tested were 40 nM, 80 nM, and 200 nM. Assays monitoring ubiquitination of *EMI1DLZC in the presence of a truncation of HSL1 (residues 768–842) from Saccharomyces cerevisiae or a UBE2SCTP (KKLAAKKKTDKKRALRRL) were conducted by titrating HSL1 (0.2, 0.5, 2, 5 μM) or UBE2SCTP (1, 2.5, 10, 25 μM). Reactions were quenched after 30 min (Figure 2e).
4.3. Cell extract experiments
HeLa S3 extracts were prepared similar to previous studies. 40 , 53 , 54 For degradation assays, extracts were used at 1:2 ratio, diluted with SB buffer (20 mM HEPES pH 7.5, 1.5 mM MgCl2, 1 mM DTT, 5 mM KCl, 2 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM AEBSF), 1 mg/ml ubiquitin (final), and energy mix (375 mM creatine phosphate, 50 mM ATP, and 50 mM MgCl2, pH 8.0). Flag‐EMI1WT‐SKP1 or EMI1NFDLZ‐SPK1 was added at 0.1 μM final concentration before reactions were allowed to procced at 30°C. At the indicated time points, aliquots of the reaction were taken and quenched in 4× SDS loading buffer before being boiled for 5 min and separated on SDS‐PAGE gels (Bio‐Rad). Proteins were transferred to nitrocellulose membranes (Bio‐Rad) and blocked for 1 hr in PBS with 0.5% Tween‐20 (PBS‐T) and 5% non‐fat dry milk (Securin, Vinculin, CDC27) or overnight (Flag, Cyclin B) before being washed three times with PBS‐T and being incubated overnight with indicated antibodies in PBS‐T with 5% BSA. Flag and Cyclin B primary antibodies were incubated for 1 hr at RT, then all membranes were washed with PBS‐T and incubated with appropriate secondary antibodies for 1 hr at room temperature. After being washed twice more with PBS‐T, membranes were incubated with ECL (Clarity™, Bio‐Rad) for 5 min and visualized on film. Antibody information: Vinculin (Santa Cruz Biotechnology, Cat. #sc‐25336, 1:1000 dilution), CyclinB (Abcam, Cat. #ab32053, 1:10,000 dilution), Securin (Santa Cruz Biotechnology. Cat. #sc‐56207, 1:1000 dilution), and Flag (Sigma Aldrich, Cat. # F3165, 1:10,000 dilution).
CONFLICT OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Derek L. Bolhuis: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); validation (equal); writing – original draft (equal); writing – review and editing (equal). Raquel C. Martinez‐Chacin: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); resources (equal); writing – original draft (equal); writing – review and editing (equal). Kaeli A. Welsh: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); resources (equal); validation (equal); writing – original draft (equal); writing – review and editing (equal). Tatyana Bodrug: Resources (supporting). Liying Cui: Resources (supporting). Michael J. Emanuele: Funding acquisition (supporting); project administration (supporting); writing – review and editing (supporting). Nicholas G. Brown: Conceptualization (lead); formal analysis (lead); funding acquisition (lead); supervision (equal); writing – original draft (lead); writing – review and editing (lead).
ACKNOWLEDGEMENTS
Our work is supported by NIH T32GM008570 (Derek L. Bolhuis and Tatyana Bodrug) and NSF DGE‐1650116 (Tatyana Bodrug); NIH T32GM135095 (Kaeli A. Welsh); NIH P30CA016086 (UNC High‐throughput peptide Synthesis Facility and Array Facility); NIH R01GM120309 and the American Cancer Society RSG‐18‐220‐01‐TBG (Michael J. Emanuele); and NIH R35GM128855 and UCRF (Nicholas G. Brown).
Bolhuis DL, Martinez‐Chacin RC, Welsh KA, Bodrug T, Cui L, Emanuele MJ, et al. Examining the mechanistic relationship of APC/CCDH1 and its interphase inhibitor EMI1 . Protein Science. 2022;31(6):e4324. 10.1002/pro.4324
Derek L. Bolhuis, Raquel C. Martinez‐Chacin, and Kaeli A. Welsh contributed equally to this study.
Review Editor: John Kuriyan
Funding information American Cancer Society, Grant/Award Number: RSG‐18‐220‐01‐TBG; National Cancer Institute, Grant/Award Number: P30CA016086; National Institute of General Medical Sciences, Grant/Award Numbers: R01GM120309, R35GM128855, T32GM008570, T32GM135095; National Science Foundation, Grant/Award Number: DGE‐1650116
REFERENCES
- 1. King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274(5293):1652–1659. [DOI] [PubMed] [Google Scholar]
- 2. Peters JM. Scf and apc: The yin and yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10(6):759–768. [DOI] [PubMed] [Google Scholar]
- 3. Visintin R, Prinz S, Amon A. Cdc20 and cdh1: A family of substrate‐specific activators of apc‐dependent proteolysis. Science. 1997;278(5337):460–463. [DOI] [PubMed] [Google Scholar]
- 4. Rape M. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol. 2018;19(1):59–70. [DOI] [PubMed] [Google Scholar]
- 5. Paiva SL, Crews CM. Targeted protein degradation: Elements of protac design. Curr Opin Chem Biol. 2019;50:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alfieri C, Zhang S, Barford D. Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (apc/c). Open Biol. 2017;7(11):1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bodrug T, Welsh KA, Hinkle M, Emanuele MJ, Brown NG. Intricate regulatory mechanisms of the anaphase‐promoting complex/cyclosome and its role in chromatin regulation. Front Cell Dev Biol. 2021;9:687515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin‐conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6(4):455–466. [DOI] [PubMed] [Google Scholar]
- 9. Yamano H, Gannon J, Hunt T. The role of proteolysis in cell cycle progression in schizosaccharomyces pombe. EMBO J. 1996;15(19):5268–5279. [PMC free article] [PubMed] [Google Scholar]
- 10. Aristarkhov A, Eytan E, Moghe A, Admon A, Hershko A, Ruderman JV. E2‐c, a cyclin‐selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc Natl Acad Sci U S A. 1996;93(9):4294–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20s complex containing cdc27 and cdc16 catalyzes the mitosis‐specific conjugation of ubiquitin to cyclin b. Cell. 1995;81(2):279–288. [DOI] [PubMed] [Google Scholar]
- 12. Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. Cdc27hs colocalizes with cdc16hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81(2):261–268. [DOI] [PubMed] [Google Scholar]
- 13. Sudakin V, Ganoth D, Dahan A, et al. The cyclosome, a large complex containing cyclin‐selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6(2):185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for b‐type cyclin proteolysis in budding yeast. Cell. 1995;81(2):269–278. [DOI] [PubMed] [Google Scholar]
- 15. Lukas C, Sorensen CS, Kramer E, et al. Accumulation of cyclin b1 requires e2f and cyclin‐a‐dependent rearrangement of the anaphase‐promoting complex. Nature. 1999;401(6755):815–818. [DOI] [PubMed] [Google Scholar]
- 16. Choudhury R, Bonacci T, Arceci A, et al. Apc/c and scf(cyclin f) constitute a reciprocal feedback circuit controlling s‐phase entry. Cell Rep. 2016;16(12):3359–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cappell SD, Chung M, Jaimovich A, Spencer SL, Meyer T. Irreversible apc(cdh1) inactivation underlies the point of no return for cell‐cycle entry. Cell. 2016;166(1):167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eguren M, Manchado E, Malumbres M. Non‐mitotic functions of the anaphase‐promoting complex. Semin Cell Dev Biol. 2011;22(6):572–578. [DOI] [PubMed] [Google Scholar]
- 19. Huang J, Bonni A. A decade of the anaphase‐promoting complex in the nervous system. Genes Dev. 2016;30(6):622–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kernan J, Bonacci T, Emanuele MJ. Who guards the guardian? Mechanisms that restrain apc/c during the cell cycle. Biochim Biophys Acta Mol Cell Res. 2018;1865(12):1924–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the apc/c and its mechanism of protein ubiquitination. Nature. 2015;522(7557):450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frye JJ, Brown NG, Petzold G, et al. Electron microscopy structure of human apc/c(cdh1)‐emi1 reveals multimodal mechanism of e3 ligase shutdown. Nat Struct Mol Biol. 2013;20(7):827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105(5):645–655. [DOI] [PubMed] [Google Scholar]
- 24. Hsu JY, Reimann JD, Sorensen CS, Lukas J, Jackson PK. E2f‐dependent accumulation of hemi1 regulates s phase entry by inhibiting apc(cdh1). Nat Cell Biol. 2002;4(5):358–366. [DOI] [PubMed] [Google Scholar]
- 25. Wang W, Kirschner MW. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase‐promoting complex. Nat Cell Biol. 2013;15(7):797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cappell SD, Mark KG, Garbett D, Pack LR, Rape M, Meyer T. Emi1 switches from being a substrate to an inhibitor of apc/c(cdh1) to start the cell cycle. Nature. 2018;558(7709):313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deshaies RJ, Joazeiro CA. Ring domain e3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. [DOI] [PubMed] [Google Scholar]
- 28. Baek K, Scott DC, Schulman BA. Nedd8 and ubiquitin ligation by cullin‐ring e3 ligases. Curr Opin Struct Biol. 2020;67:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davey NE, Morgan DO. Building a regulatory network with short linear sequence motifs: Lessons from the degrons of the anaphase‐promoting complex. Mol Cell. 2016;64(1):12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. da Fonseca PC, Kong EH, Zhang Z, et al. Structures of apc/c(cdh1) with substrates identify cdh1 and apc10 as the d‐box co‐receptor. Nature. 2011;470(7333):274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buschhorn BA, Petzold G, Galova M, et al. Substrate binding on the apc/c occurs between the coactivator cdh1 and the processivity factor doc1. Nat Struct Mol Biol. 2011;18(1):6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang LF, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase‐promoting complex. Nature. 2014;513(7518):388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garnett MJ, Mansfeld J, Godwin C, et al. Ube2s elongates ubiquitin chains on apc/c substrates to promote mitotic exit. Nat Cell Biol. 2009;11(11):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological e2 module for the human anaphase‐promoting complex. Proc Natl Acad Sci USA. 2009;106(43):18213–18218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. Ube2s drives elongation of k11‐linked ubiquitin chains by the anaphase‐promoting complex. Proc Natl Acad Sci USA. 2010;107(4):1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage‐specific ubiquitin chain elongation by a single‐subunit e2. Cell. 2011;144(5):769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown NG, VanderLinden R, Watson ER, et al. Dual ring e3 architectures regulate multiubiquitination and ubiquitin chain elongation by apc/c. Cell. 2016;165(6):1440–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brown NG, VanderLinden R, Watson ER, et al. Ring e3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase‐promoting complex. Proc Natl Acad Sci USA. 2015;112(17):5272–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamaguchi M, VanderLinden R, Weissmann F, et al. Cryo‐em of mitotic checkpoint complex‐bound apc/c reveals reciprocal and conformational regulation of ubiquitin ligation. Mol Cell. 2016;63(4):593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martinez‐Chacin RC, Bodrug T, Bolhuis DL, et al. Ubiquitin chain‐elongating enzyme ube2s activates the ring e3 ligase apc/c for substrate priming. Nat Struct Mol Biol. 2020;27(6):550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reimann JD, Gardner BE, Margottin‐Goguet F, Jackson PK. Emi1 regulates the anaphase‐promoting complex by a different mechanism than mad2 proteins. Genes Dev. 2001;15(24):3278–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miller JJ, Summers MK, Hansen DV, et al. Emi1 stably binds and inhibits the anaphase‐promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 2006;20(17):2410–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Q, Chang L, Aibara S, Yang J, Zhang Z, Barford D. Wd40 domain of apc1 is critical for the coactivator‐induced allosteric transition that stimulates apc/c catalytic activity. Proc Natl Acad Sci USA. 2016;113(38):10547–10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moustafa D, Elwahed MRA, Elsaid HH, Parvin JD. Modulation of early mitotic inhibitor 1 (emi1) depletion on the sensitivity of parp inhibitors in brca1 mutated triple‐negative breast cancer cells. PLoS One. 2021;16(1):e0235025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marzio A, Puccini J, Kwon Y, et al. The f‐box domain‐dependent activity of emi1 regulates parpi sensitivity in triple‐negative breast cancers. Mol Cell. 2019;73(2):224–237 e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vaidyanathan S, Cato K, Tang L, et al. In vivo overexpression of emi1 promotes chromosome instability and tumorigenesis. Oncogene. 2016;35(41):5446–5455. [DOI] [PubMed] [Google Scholar]
- 47. Guan C, Zhang J, Zhang J, Shi H, Ni R. Enhanced expression of early mitotic inhibitor‐1 predicts a poor prognosis in esophageal squamous cell carcinoma patients. Oncol Lett. 2016;12(1):114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rubin SM, Sage J, Skotheim JM. Integrating old and new paradigms of g1/s control. Mol Cell. 2020;80(2):183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Emanuele MJ, Enrico TP, Mouery RD, Wasserman D, Nachum S, Tzur A. Complex cartography: Regulation of e2f transcription factors by cyclin f and ubiquitin. Trends Cell Biol. 2020;30(8):640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sadasivam S, DeCaprio JA. The dream complex: Master coordinator of cell cycle‐dependent gene expression. Nat Rev Cancer. 2013;13(8):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lehman NL, Tibshirani R, Hsu JY, et al. Oncogenic regulators and substrates of the anaphase promoting complex/cyclosome are frequently overexpressed in malignant tumors. Am J Pathol. 2007;170(5):1793–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eldridge AG, Loktev AV, Hansen DV, Verschuren EW, Reimann JD, Jackson PK. The evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase‐promoting complex inhibitor emi1. Cell. 2006;124(2):367–380. [DOI] [PubMed] [Google Scholar]
- 53. Williamson A, Jin L, Rape M. Preparation of synchronized human cell extracts to study ubiquitination and degradation. Methods Mol Biol. 2009;545:301–312. [DOI] [PubMed] [Google Scholar]
- 54. Welsh KA, Bolhuis DL, Nederstigt AE, et al. Functional conservation and divergence of the helix‐turn‐helix motif of e2 ubiquitin‐conjugating enzymes. EMBO J. 2022;41(3):e108823. [DOI] [PMC free article] [PubMed] [Google Scholar]


