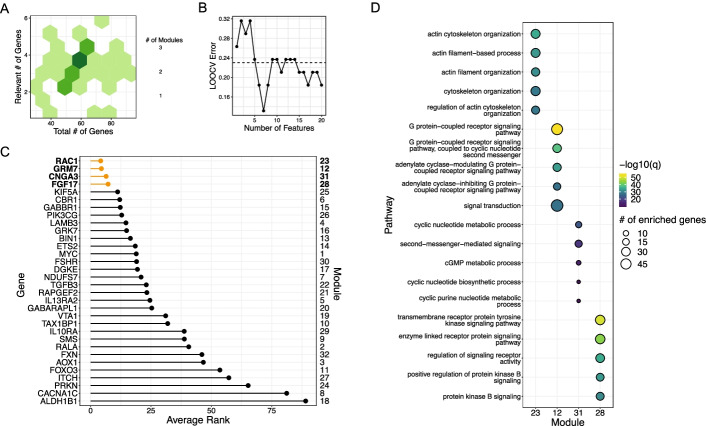
Fig. 3.
Pathways that inform BRAFi response. A Visualization of gene distribution within modules. Relevant genes are those that passed the Boruta filtering step. X-axis denote total number genes per module, y-axis denotes number of relevant genes, and the shading indicates total number of modules. B Leave-one-out cross validation error as a function of number of features used in the SVM model. Dashed line indicates no information rate, i.e. the error made if the class with the greatest frequency was selected. C Minimum feature ranking for each module. D GO Biological Processes pathway enrichment of genes contained within modules presented in C). P-values shown are corrected for multiple hypothesis testing using the Holm-Bonferroni method