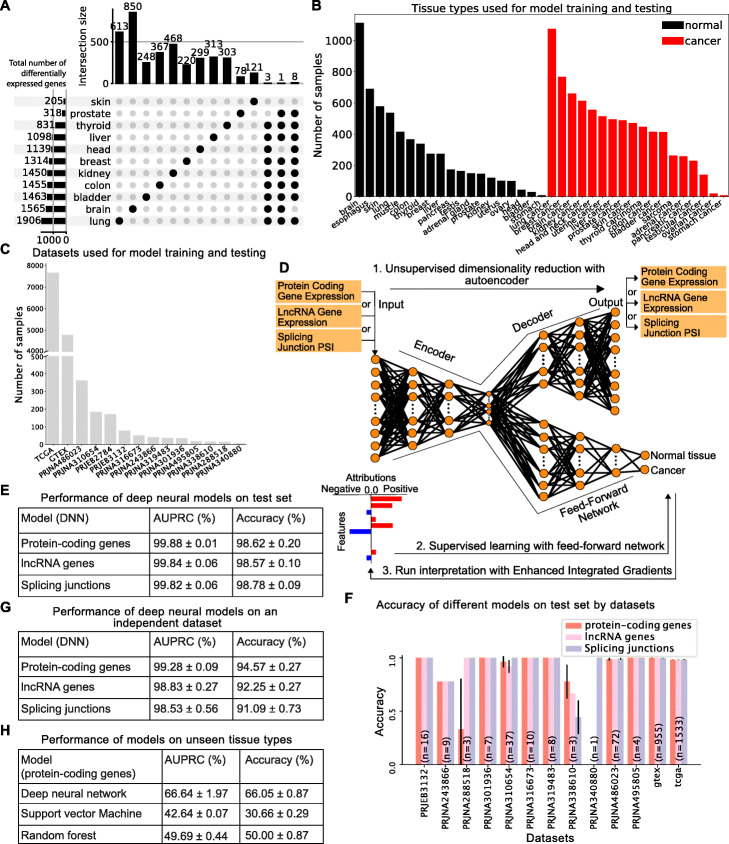
Fig. 1.
A Upset plot summarizing pairwise differential gene expression analyses performed on tumors and their corresponding normal tissue. No gene is significantly deregulated in more than 9 out of 11 cancer types tested. B, C Dataset assembled to train and test binary models to distinguish between normal and tumor samples shown by tissue type (B) or dataset (C). D Graphical representation of the computational framework used to train, test, and interpret the models. E Performance of models trained with protein-coding gene expression, lncRNA gene expression, or splicing variations evaluated by area under the precision-recall curve (AUPRC) and accuracy (sum of true positives and true negatives over the total population). F Accuracy of models trained with protein-coding gene expression, lncRNA gene expression, or splice junction usage across the 13 datasets used to assemble the training set. G Performance of models trained with protein-coding gene expression, lncRNA gene expression, or splice junction usage on an independent dataset consisting of normal and cancer lung samples. H Performance of the deep learning model, SVM, and random forest using protein-coding gene expression on unseen tissue types (blood cancers) with no batch correction. The training set consists of solid tumors only