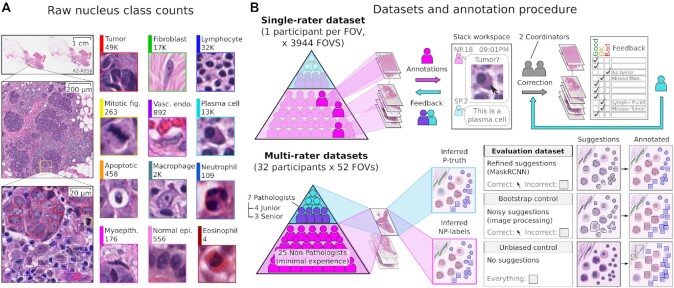
Figure 1:
Dataset annotation and quality control procedure. A. Nucleus classes annotated. B. Annotation procedure and resulting datasets. Two approaches were used to obtain nucleus labels from non-pathologists (NPs). (Top) The first approach focused on breadth, collecting single-rater annotations over a large number of FOVs to obtain the majority of data in this study. NPs were given feedback on their annotations, and 2 study coordinators corrected and standardized all single-rater NP annotations on the basis of input from a senior pathologist. (Bottom) The second approach evaluated interrater reliability and agreement, obtaining annotations from multiple NPs for a smaller set of shared FOVs. Annotations were also obtained from pathologists for these FOVs to measure NP reliability. The procedure for inferring a single set of labels from multiple participants is described in Fig. 2. We distinguished between inferred non-pathologist labels (NP-labels) and inferred pathologist truth (P-truth) for clarity. Three multi-rater datasets were obtained: an Evaluation dataset, which is the primary multi-rater dataset, as well as Bootstrap and Unbiased experimental controls to measure the value of algorithmic suggestions. In all datasets except the Unbiased control, participants were shown algorithmic suggestions for nucleus boundaries and classes. They were directed to click nuclei with correct boundary suggestions and annotate other nuclei with bounding boxes. The pipeline to obtain algorithmic suggestions consisted of 2 steps: (i) Using image processing to obtain bootstrapped suggestions (Bootstrap control); (ii) Training a Mask R-CNN deep-learning model to refine the bootstrapped suggestions (single-rater and Evaluation datasets).