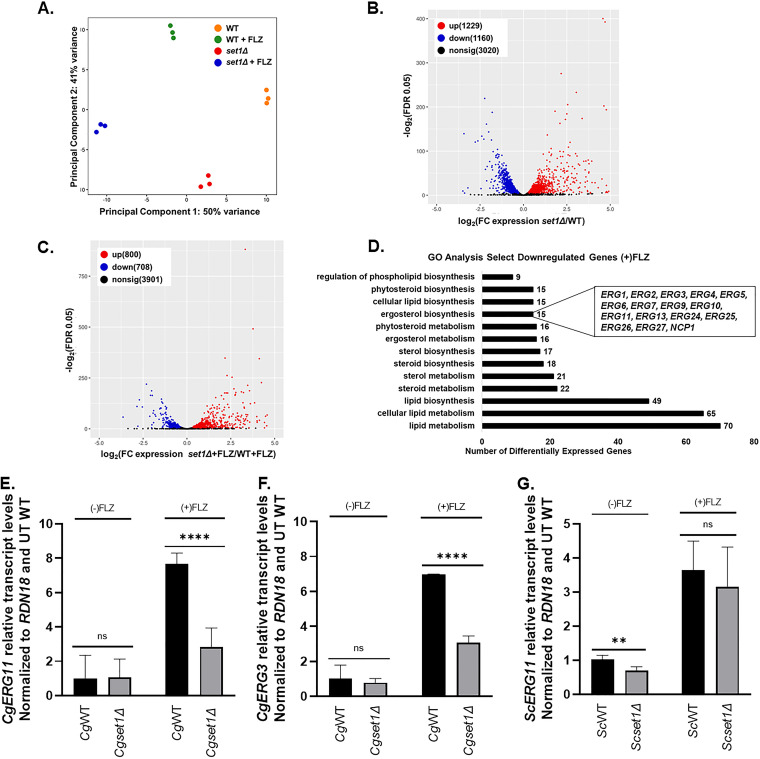
FIG 5.
The deletion of SET1 in C. glabrata alters global and local levels of gene expression under untreated and azole-treated conditions. The genomewide changes in gene expression under azoles were assessed using C. glabrata CBS138 (2001) WT and set1Δ strains. (A) PCA for WT and set1Δ azole-treated samples relative to WT untreated samples based on the counts per million. (B) Volcano plot showing the significance [−log2(FDR), y axis] versus the fold change (x axis) of the DEGs identified in the WT untreated samples relative to set1Δ untreated samples. (C) Volcano plot showing the significance [−log2(FDR), y axis] versus the fold change (x axis) of the DEGs identified in the set1Δ azole-treated samples relative to WT azole-treated samples. Genes with significant differential expression (FDR < 0.05) in panels B and C are highlighted in red and blue for up- and downregulated genes, respectively. Black highlighted genes are considered nonsignificant. (D) Genes from the RNA-seq data set that were statistically significantly enriched (FDR < 0.05) were used for GO term determination of Set1-dependent DEGs under azole conditions. Downregulated genes refer to the DEGs that are dependent on Set1 for activation either directly or indirectly. Significantly enriched groups of GO terms were identified as the DEGs from only set1Δ and WT azole-treated samples. (E to G) Expression of indicated genes was determined in WT and set1Δ strain cells in either C. glabrata or S. cerevisiae treated with 64 μg/mL fluconazole for 3 h by qRT-PCR analysis. Gene expression analysis was set relative to the untreated wild-type and expression was normalized to RDN18 mRNA levels. Data were analyzed from ≥3 biological replicates with three technical replicates each. Statistics were determined using the GraphPad Prism Student t test, version 9.2.0. ****, P < 0.0001; **, P < 0.01; ns, P > 0.05. Error bars represent the SD.