Figure 2. Overview of HCS workflows.
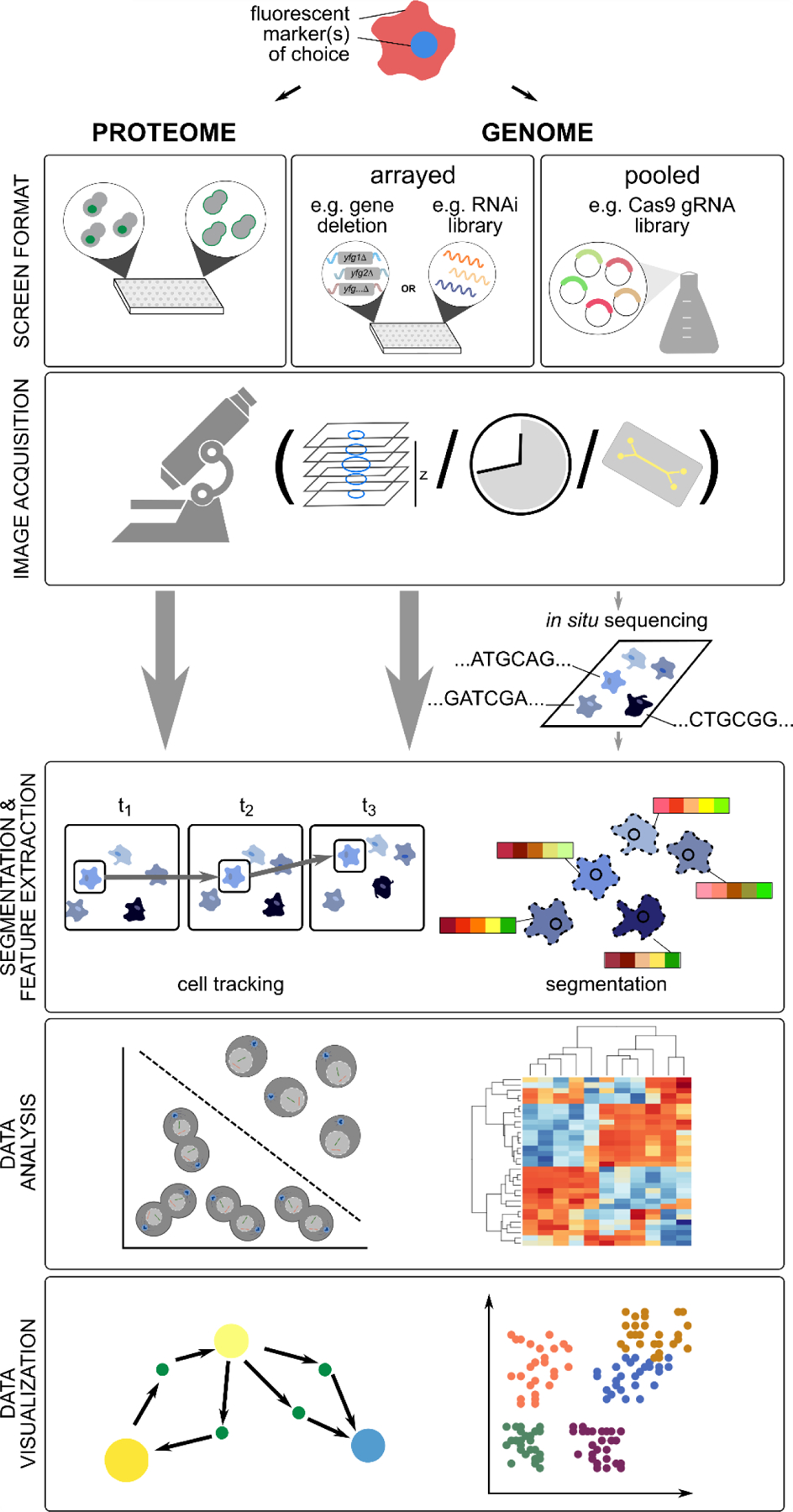
HCS screens, both to monitor proteome-wide changes and large-scale genetic perturbations, are generally performed in arrayed format, but more recently pooled formats have been used. Image acquisition (high-throughput live or fixed-cell imaging of fluorescent markers) may include multiple planes and/or time points. Microfluidics devices may be used to track changes after switching growth conditions or to capture and monitor a subset of cells. Pooled screening approaches require post-imaging deconvolution (e.g. with in situ sequencing) to identify the underlying perturbation. After image acquisition, individual cells are segmented, and in case of time-lapse imaging, tracked, based on positional information. Numeric features describing different cell/phenotype properties are extracted, and the derived feature vectors analyzed with different machine learning approaches, including classification and clustering. Network analysis and feature projection techniques are often used for data visualization. See main text for more details.
