| |
|---|---|
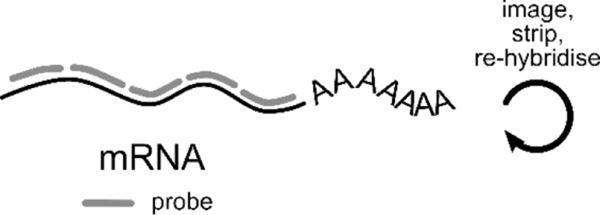
| Fluorescence in situ hybridisation (FISH) of RNA offers spatial information unobtainable from sequencing. It is the gold standard for accurately determining RNA counts and can capture up to 10000 unique transcripts (Torre et al., 2018). Through iterative rounds of imaging, hybridisation, and re-probing (cartoon above), RNA species are identified by different imaging readouts (cartoons below). Some recent single molecule FISH (smFISH) technologies include: | |
readout

|
osmFISH (cyclic-ouroboros
smFISH) The simplest type of FISH, osmFISH, is a non-barcoded strategy that has a high dynamic range. In each cycle, each DNA probe labelled with a unique fluorophore targets a distinct mRNA. The number of targets detected is the product of the number of fluorescent channels and the number of cycles. This technique was used to map the somatosensory cortex in mice (Codeluppi et al., 2018). |
| The remaining approaches we outline here require multiple cycles to identify each RNA. | |
|
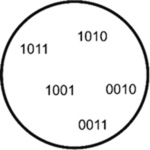
MERFISH (Multiplexed
error-robust FISH) MERFISH assigns 2-bit barcodes (0 for absence, 1 for presence of fluorescence) to RNA species through iterative rounds of hybridization and imaging (Moffitt et al., 2016). This way, in 15 cycles, up to 2^15 (~30000) unique barcodes can be assigned. However, to allow for error correction, some barcodes are left intentionally unassigned. MERFISH was used to identify ~1600 cell-cycle dependent genes as well as RNAs enriched in the different compartments in U-2 OS cells (Xia et al., 2019). |
|
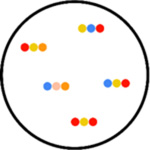
STARmap (spatially resolved
transcript amplicon readout mapping) STARmap is useful for 3-D systems. RNA species are paired with DNA probes in order to produce a DNA nanoball using amplification. Here, hydrogel-tissue chemistry is used to preserve the spatial relationships in tissues. Fluorescent-based sequence readout is then used to identify RNA transcripts. STARmap was used in the mapping of 1000 genes of the medial prefrontal cortex of the adult mouse brain (Wang et al., 2018). |
|
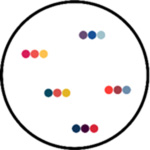
seqFISH+ (sequential
fluorescent in situ
hybridisation) seqFISH+ uses super-resolution confocal microscopy to avoid optical crowding, and pseudo-colors to reduce imaging cycles. Each mRNA species has a unique color sequence, so the number of unique barcodes assigned is F^n (F=fluorophores, n=cycles). seqFISH(+) was used to characterize the transcriptomes of tissues such as the cortex, sub-ventricular zone and olfactory bulb in mice and to capture spatial information between ligand-receptor pairs in neighboring cells (Eng et al., 2019; Eng et al., 2017). |

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.