Abstract
Objectives.
Asthma is a heterogenous condition with significant diagnostic complexity, including variations in symptoms and temporal criteria. The disease can be difficult for clinicians to diagnose accurately. Properly identifying asthma patients from the electronic health record is consequently challenging as current algorithms (computable phenotypes) rely on diagnostic codes (e.g., International Classification of Disease, ICD) in addition to other criteria (e.g., inhaler medications) - but presume an accurate diagnosis. As such, there is no universally accepted or rigorously tested computable phenotype for asthma.
Methods.
We compared two established asthma computable phenotypes: the Chicago Area Patient-Outcomes Research Network (CAPriCORN) and Phenotype KnowledgeBase (PheKB). We established a large-scale, consensus gold standard (n=1,365) from the University of California, Los Angeles Health System’s clinical data warehouse for patients 5–17 years old. Results were manually reviewed and predictive performance (positive predictive value, sensitivity/specificity, F1-score) determined. We then examined the classification errors to gain insight for future algorithm optimizations.
Results.
As applied to our final cohort of 1,365 expert-defined gold standard patients, the CAPriCORN algorithms performed with a balanced positive predictive value (PPV)=95.8% (95% CI: 94.4–97.2%), sensitivity=85.7% (95% CI: 83.9–87.5%), and harmonized F1=90.4% (95% CI: 89.2–91.7%). The PheKB algorithm performed with a balanced PPV=83.1% (95% CI: 80.5–85.7%), sensitivity=69.4% (95% CI: 66.3–72.5%), and F1=75.4% (95% CI: 73.1–77.8%). Four categories of errors were identified related to method limitations, disease definition, human error, and design implementation.
Conclusions.
The performance of the CAPriCORN and PheKB algorithms was lower than previously reported as applied to pediatric data (PPV=97.7% and 96%, respectively). There is room to improve the performance of current methods, including targeted use of natural language processing and clinical feature engineering.
Keywords: Asthma, Computable Phenotype, Data Standardization, Electronic Health Records, Pediatrics
INTRODUCTION
Cohort discovery and identification of patients from the electronic health record (EHR) is often achieved by implementing a “computable phenotype,” defined by a set of inclusion and exclusion rules involving structured clinical codes and observations (e.g., disease codes, medications, keywords, lab values, etc.) that are collectively indicative of a target disease.1 Computable phenotypes representing medical conditions have been developed for a number of diseases, including diabetes and obesity.2,3 However, there is not yet a universal approach for development of these methods.4 In addition, there is no central repository, with the algorithms found among several platforms including the Phenotype Knowledgebase (PheKB),4 the National Committee for Quality Assurance (NCQA) through the Healthcare Effectiveness Data and Information Set (HEDIS),5 the Centers for Medicare and Medicaid Services,6 the Agency for Healthcare Research and Quality (AHRQ),7 the National Quality Forum,8 the phenotype execution and modeling architecture (PhEMA),9 the National Patient-Centered Clinical Research Network (PCORnet),10 the Observational Health Data Sciences and Informatics (OHDSI),11 and individual academic institutional publications.12–14 Markedly, computational phenotyping objectives can vary in intent, and it is often unclear for what purpose it was optimized, with no common standards around the documentation or evaluation of these methods. For example, cohort identification purpose can range from use in research case-control to revenue cycle inquiries and quality improvement measures. Consequently, it can be difficult for end users to decide which computable phenotype to use for a specific need or if one is generalizable to their population of interest. As a result, the application of these algorithms are inconsistent or non-generalizable across different tasks or patient populations, and performance may not match expectations.15–19
To help understand the complexities and performance of computational phenotypes, we chose to study pediatric asthma. In the United States, asthma is one of the most common chronic illnesses of childhood.20 Many factors contribute to its presentation and symptom control including variable underlying inflammation, medication response, and socioeconomic and environmental stressors, where it leads to high rates of hospitalizations, healthcare costs, and school absenteeism.17,21 Yet despite its prevalence, pediatric asthma remains difficult to diagnose and there is increasing attention on the use of the EHR both to identify untreated individuals, as well as to elucidate the condition and derive new insights to better personalize treatment outcomes.22 With its phenotypic complexity, symptoms that overlap with similar diseases as well as differences in diagnosis and clinical treatment across institutions makes a consistent, rule-based definition of pediatric asthma difficult to establish.
We evaluated two computable phenotypes: 1) the Chicago-based CAPriCORN Asthma Cohort Committee algorithm12,23 and 2) a PheKB algorithm, developed at the Children’s Hospital of Philadelphia (CHOP).24 Both methods are published and report statistical results for identification of asthma patients from institutional EHRs. Each were motivated by different purposes - the PheKB algorithm for identification of asthma cases in a genome-wide association research study, while the CAPriCORN algorithm was validated for generalizability to identify asthma cases across Chicago-area institutions. We tested these methods on a large, UCLA-derived cohort of pediatric patients to ascertain predictive performance as well as to gain deeper insights into the difficulties of reproducing computational phenotyping approaches.
OBJECTIVES
Our goals were to assess and compare the performance of the CAPriCORN and PheKB asthma computable phenotype algorithms on a subset of pediatric patients drawn from the UCLA Health System population. We specifically examined the error types that arise in these asthma computable phenotypes as part of the design requirement to propose steps for rule optimization.
METHODS
Healthcare system
The study was conducted within the University of California Los Angeles (UCLA) Health System, an academic quaternary care facility. UCLA Health System includes four hospitals and over 180 outpatient clinics across the Los Angeles area. It has a catchment area of over 4 million people with a wide range of socioeconomic status, cultures, and payor mix. Our inclusion criteria were children 5–17 years old as of February 28, 2019 with more than one encounter at any time within the UCLA Health System’s electronic health record (Epic Systems) data warehouse.
Computable phenotypes
The CAPriCORN algorithm was developed at the University of Chicago by Pacheco et al. and later modified by Afshar et al. with the goal of identifying asthma patient cohorts for research within the Patient-Centered Outcomes Research (PCORI) Clinical Data Research Network.12,23 The computable phenotype was applied to a population ages 5–89 years old. The PheKB algorithm was developed by Almoguera et al. at CHOP within a study to identify genetic markers in asthma patients and the average age of participants was 11-years old.24
Details of the CAPriCORN and PheKB algorithms are depicted in Table 1. We modified the inclusion criteria to include only children ages 5–17 years old and included ICD-9 codes as well as mappings to their equivalent ICD-10 diagnosis codes. To map ICD-9 to ICD-10 codes, we used the General Equivalence Mappings (GEMS) data method published by the Centers for Medicare and Medicaid Services (CMS).25
Table 1.
Rules derived from the CAPriCORN and PheKB algorithms to identify patients with asthma as applied to the UCLA Health System Clinical Data Warehouse.
| CAPriCORN | PheKB |
|---|---|
| At least 2 encounters | At least 2 encounters |
| Ages 5–17 years old | Ages 5–17 years old |
|
Visit 1: Asthma-related diagnosis AND Visit 2: Asthma-related diagnosis OR an asthma medicationa |
Visit 1 &2: ≥1 asthma-related medicationa AND diagnosis OR ≥=3 visits in 1 year (separate calendar days) with “wheezing” or “asthma” documented in the note |
| No exclusion criteriab | No exclusion criteriab |
Asthma-related diagnosis = ICD-9 code 493.* and/or ICD-10 code J45.*
Asthma medication detail in Appendix Table 2
Exclusion criteria detail in Appendix Table 3
Gold standard creation
We performed our analysis on a subset of manually reviewed patients (i.e., the gold standard), which were drawn from the UCLA pediatric patient population, ages 517. We defined true asthma status (positive or negative) based on the National Institutes of Health National Asthma Education and Prevention Program (NAEPP) and the Global Initiative for asthma (GINA) guideline definitions of asthma (Appendix, Table 1).17,26 Case review using this definition guide was performed by pediatric residents and pre-medical students (M.R., A.N., H.H., and A.L.) and all cases were reviewed and adjudicated by a pediatric pulmonologist (M.K.R.). A set of 500 previously identified possible asthma cases was then matched with a set of 500 predicted negative controls as defined by CAPriCORN exclusion criteria and a set of 500 predicted negative controls as defined by PheKB exclusion criteria, for a set of n=1,500 patients. After removal of duplicates, cases were classified by the authors into three categories: asthma positive, asthma negative, or ambiguous. The ambiguous category consisted of cases classified as “probable asthma” or “possible asthma.” “Probable” cases contained either diagnoses and/or medication related to asthma without documented confirmation of clinical treatment response, pulmonary function testing, etc. “Possible” cases were those with an asthma diagnosis documented in the past medical history or problem list but without supporting clinical documentation. Only non-ambiguous cases were used for the final analysis, yielding 1,365 cases.
Algorithm evaluation
We extracted information from the clinical data warehouse needed to execute the CAPriCORN and PheKB computable phenotype rules (patient encounters, demographics, visit diagnoses, clinical text, and medications), applied the algorithms to predict a label for each child of being asthma “positive” or “negative,” and matched the labeled cases to the gold standard. We calculated positive and negative predictive values (PPV, NPV), sensitivity, specificity, and F1-score of each computable phenotype. Agreement between the CAPriCORN and PheKB algorithms to identify asthma cases was evaluated using Cohen’s Kappa score. The positive cases (n=375) were upsampled to match the n=990 control patients and test performance in a balanced manner. Twenty subsamples of 200 patients were used to bootstrap confidence intervals of performance metrics. Discrepancies between the algorithms’ determination of asthma status were quantified. To reflect PheKB’s and CAPriCORN’s performance (which were originally evaluated on balanced datasets), we reviewed performance with the original distribution of 375/990 cases/controls as well as after balancing.
To further understand algorithm differences, the false positives and false negative cases were manually reviewed by the domain expert (M.K.R.) and categorized into themes based on best judgment to describe the reason for error. We also reviewed the specific computational phenotype rules, the corresponding potential to cause incorrect classification, and potential solutions.
Ethical considerations
A waiver of consent was obtained from the UCLA Institutional Review Board (IRB) committee (IRB #18–002015).
RESULTS
100,869 pediatric patients aged 5–17 met our initial inclusion criteria, and from this cohort, the CAPriCORN and PheKB algorithms were separately applied to build the gold standard. After manual review, the ambiguous asthma status and overlapping cases were excluded for a final gold standard set of 1,365 children in our analysis: 375 children (27%) with asthma and 990 without (73%), demographics Table 2.
Table 2.
Demographics of pediatric asthma patients included in the study
| Demographics | Total (n=1365) | Asthma (n=375) | No Asthma (n=990) | p-value |
|---|---|---|---|---|
| Age (avg/sd) | 11.9 / 3.86 | 11.43 / 3.69 | 11.37 / 3.93 | |
| Female | 623 | 148 | 475 | 0.1 15 |
| Male | 742 | 227 | 515 | 0.163 |
| Race | ||||
| Asian | 111 | 44 | 67 | 0.016 |
| Black | 77 | 33 | 44 | 0.010 |
| White | 621 | 170 | 451 | 0.998 |
| Other | 556 | 128 | 428 | 0.063 |
| Ethnicity | ||||
| Hispanic/Latino(a) | 268 | 65 | 203 | 0.498 |
| Not Hispanic/Latino(a) | 775 | 245 | 530 | 0.035 |
| Unknown | 322 | 65 | 257 | 0.013 |
Performance
Applied to pediatric patients within the UCLA Health System, the unbalanced CAPriCORN rule’s performance resulted in a PPV of 90.0% (95% CI: 85.5–92.3%), sensitivity of 89.0% (95% CI: 82.7–90.7%) and F1 of 89.5% (95% CI: 84.7–90.2%). The balanced CAPriCORN algorithms performed with a positive predictive value (PPV) of 95.8% (95% CI: 94.4–97.2%), sensitivity of 85.7% (95% CI: 83.9–87.5%), and F1 of 90.4% (95% CI: 89.2–91.7%); Table 3. The balanced dataset and particularly the unbalanced dataset underperformed in some areas compared with its published performance using the CAPriCORN Chicago-area data of a PPV 97.7% and sensitivity of 97.6% (n=409 children)15 and a derived F1 of 96.4%.
Table 3.
a) CAPriCORN 2×2 table (n=1365), original
| Asthma (+) | Asthma (−) | |
|---|---|---|
| CAPriCORN (+) | 334 (TP) | 37 (FP) |
| CAPriCORN (−) | 41 (FN) | 953 (TN) |
| b) CAPriCORN 2×2 table (n=1980), balanced |
| Asthma (+) | Asthma (−) | |
| CAPriCORN (+) | 867 (TP) | 37(FP) |
| CAPriCORN (−) | 123(FN) | 953 (TN) |
| c) PheKB 2×2 table (n=1365), original |
| Asthma (+) | Asthma (−) | |
| PheKB (+) | 274 (TP) | 133 (FP) |
| PheKB (−) | 101(FN) | 857 (TN) |
| d) PheKB 2×2 table (n=1980), balanced |
| Asthma (+) | Asthma (−) | |
| PheKB (+) | 687 (TP) | 133 (FP) |
| PheKB (−) | 303(FN) | 857 (TN) |
| e) PPV/NPV/Sensitivity/Specificity/F measure, original |
| N=1365 | PPV (95% CI) | NPV (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | Fl-measure |
| CAPriCORN | 90.0%(85.5%-92.3%) | 95.8%(93.3%-96.2%) | 89.0%(82.7%−90.7%) | 96.2%(94.6%−97.1%) | 89.5%(84.7%−90.2%) |
| PheKB | 67.3%(62.8%-69.8%) | 89.4%(86.3%−90.3%) | 73.0%(67.7%-75.2%) | 86.5%(83.9%−87.4%) | 70.0%(65.7%−71.0%) |
| f) PPV/NPV/Sensitivity/Specificity/F measure, balanced |
| N=1980 | PPV (95%CI) | NPV (95%CI) | Sensitivity (95% CI) | Specificity (95% CI) | Fl-measure |
| CAPriCORN | 95.8%(94.4%-97.2%) | 86.7%(85.1%−88.4%) | 85.7%(83.9%-87.5%) | 96.1%(94.8%-97.5%) | 90.4%(89.2%−91.7%) |
| PheKB | 83.1%(80.5%−85.7%) | 73.1%(70.5%−75.8%) | 69.4%(66.3%-72.5%) | 85.4%(83.3%-87.6%) | 75.4%(73.1%−77.8%) |
The unbalanced PheKB algorithm performed on the UCLA data with a PPV of 67.3% (95% CI: 62.8–69.8%), sensitivity of 73.0% (95% CI: 67.7–75.2%) and F1 of 70.0% (95% CI: 65.7–71.0%). The balanced PheKB algorithm performed with a PPV of 83.1% (95% CI: 80.5–85.7%), sensitivity of 69.4% (95% CI: 66.3–72.5%), and F1 of 75.4% (95% CI: 73.1–77.8%). The PheKB algorithm has been measured previously in terms of PPV, with a reported performance (n=25) at Marshfield Health System in Wisconsin of 96%.16 Markedly, the CAPriCORN and PheKB computable phenotype yielded a Cohen’s Kappa of 0.63 (unbalanced), and 0.60 (balanced).
Error analysis
There were four general categories of errors discovered among the computable phenotype algorithms (Table 4):
Table 4.
Categories of Error Type
| Error Category | CAPriCORN | PheKB |
|---|---|---|
| Computational | ||
| Confirmatory history in text | FN | FN |
| Scanned or outside system notes | FN | FN |
| Definition | ||
| Exclusion diagnosis missed | FP | FP |
| Diagnosis excluded unnecessarily | FN | FN |
| Only 1 respiratory episode | FP | FP |
| Diagnosis plus medication requirement | -- | FN |
| Human error | ||
| Error related to diagnosis | FN/FP | FP |
| Implementation | ||
| Text | -- | FP |
FN=false negative, FP=false positive
1. Computational.
Computational errors were those that a computable phenotype inherently cannot perform such as identifying asthma cases using details within progress notes, scanned documents, or outside records (e.g., if an outside record or scanned document contains information supportive of an asthma diagnosis, this is not captured in the algorithm rules). These are errors that arise prior to the design of a phenotyping algorithm and are difficult to perform computationally by the phenotyping algorithm itself.
2. Definition.
Examples of errors caused by the computable phenotype definition were exclusion criteria that were not identified (e.g., hypotonia) or too broad (e.g., bronchitis), only one respiratory episode counted as two episodes by the computable phenotype algorithms, and narrow criteria such as the requirement for asthma diagnosis and medication at each visit. Calibration of definitions can resolve these types of errors.
3. Human error.
If the patient was not labeled by the physician as having asthma when they met criteria or they were labeled as an asthma patient without meeting diagnostic criteria, we considered that a human diagnostic error.
4. Implementation.
The final major error type was due to the keyword rules of the PheKB computable phenotype, which only included ‘asthma’ and ‘wheezing.’ As applied to our healthcare system, the rules would require modifications such as searching roots, using ontology/concept mappings, avoiding negation, and extracting selectively within the chart to improve performance (e.g., excluding the ‘family history’ section and/or only including the ‘history of present illness’ section). More advanced tools such as advanced NLP algorithms or data warehousing can resolve these types of errors.
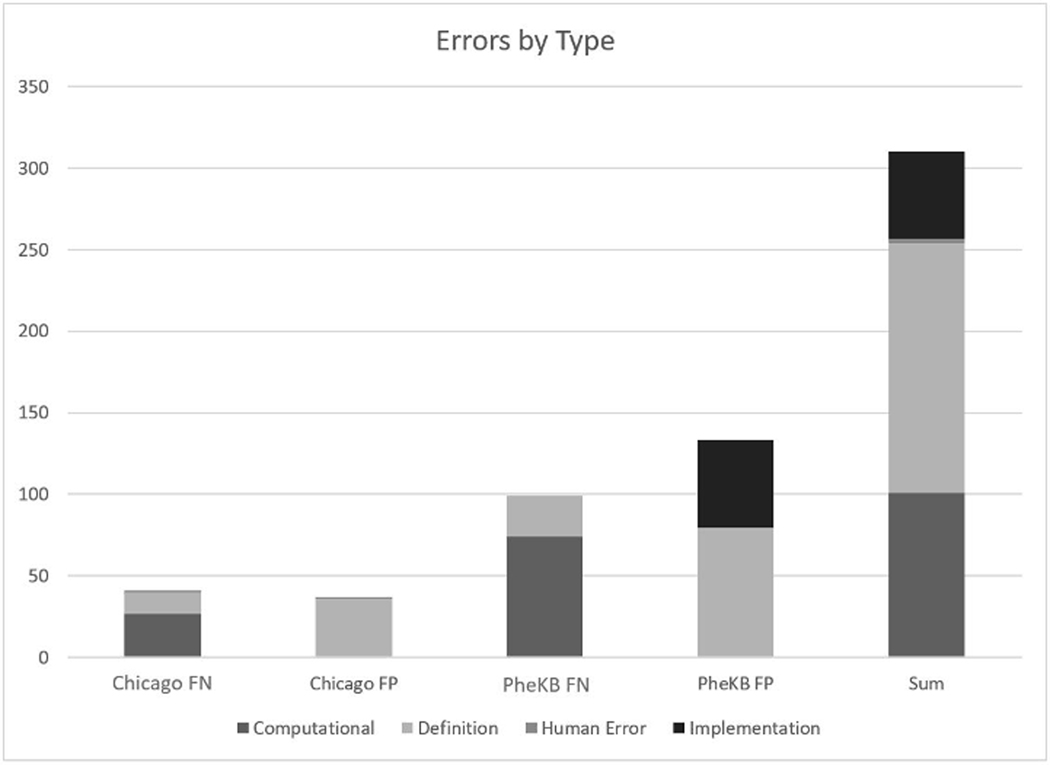
The most common type of error leading to false positive across both algorithms (and the majority of CAPriCORN’s false positive labels) were due to “definition” errors - not excluding diagnoses that could confound the presentation, diagnosis, and/or treatment of asthma (e.g., musculoskeletal disease, immune system dysfunction). The majority of the PheKB false positives were due to “implementation” errors that erroneously included text (e.g., family history of asthma). The majority of both the CAPriCORN and PheKB algorithms’ false negatives were due to information located within documentation from scanned or outside records (Figure 1).
Figure 1.
Distribution of errors by type FP=False Positive
DISCUSSION
To date, there is no singular approach to asthma case identification from the EHR; previous efforts to do so demonstrate a range of findings, with positive predictive values from 27–100%.12,23,27–35 Computable phenotypes that rely on ICD codes are vulnerable due to dependence on correct diagnosis by a healthcare provider. Our findings demonstrate the CAPriCORN and PheKB asthma computable phenotypes as applied to pediatric patients in the UCLA Health System had decreased performance. As the comparison between the PheKB and CAPriCORN algorithms demonstrate, the performance is difficult to generalize across populations and institutions. Notably, both the PheKB and CAPriCORN algorithms were designed for identifying ideal candidates for downstream asthma research by not including ambiguous cases.
While an F1-score of 90% for the CAPriCORN rule results in our system is reasonable, it is below the level observed by Pacheco et al and Afshar et al. While the CAPriCORN algorithm was designed for generalizability and validated across institutions in the Chicago area, its lower-than-expected performance on the UCLA population, particularly in terms of sensitivity, demonstrate that there is a generalization gap for the CAPriCORN algorithm. Contributing factors were likely due to a more realistic performance metric using a larger cohort of >1,000 pediatric patients, as well as institutional differences in data collection and warehousing. Arguably, these results may be acceptable depending on the application of the computable phenotype - whether recruiting, genomic-phenomic research, decision support, quality improvement, or other applications - and the need to balance true/false positive and negative rates. However, without contextualization of the computational phenotype’s primary objective and evaluation, appreciating its proper utility can be difficult.
Clinical Implications
Furthermore, the significant difference in performance of the PheKB algorithm on our population compared to the population in Pacheco et al illustrates the problems of applying algorithms at cross-purposes to their intended application. The PheKB algorithm is used as part of a wider effort to identify candidate patients for GWAS studies24. Compared to CAPriCORN, PheKB’s rules are more restrictive. The difference is demonstrated in two ways. First, PheKB’s lower sensitivity but higher specificity illustrates PheKB’s stricter rules, whereas CAPriCORN’s rules were broader to generalize across institutions with divergent clinical practice. Second, the relatively low Cohen’s Kappa between the CAPriCORN and PheKB algorithms illustrate divergence in who these algorithms consider as having asthma - CAPriCORN recognizing those patients who may only have one diagnosis documented, while PheKB uses text-based identification that does not rely on capture via ICD coding.
As part of precision medicine efforts, there is a call for standardized, automated, and portable approaches to identify patient cohorts for integration of genomic and EHR data.36 Central data repositories such as PheKB are expanding, but do not enforce standardized methods or model design principles for computable phenotype development and validation. This study demonstrates the need for more consistent thinking about the application of computable phenotypes, the different purposes that they could be used, and their issues of generalizability and transportability. In 2015, Mo et al. published a ten-component recommendation for computable phenotype representation models (PheRMs) using 21 eMERGE phenotype rules; however, asthma was not one of the computable phenotypes studied.3
Based on the error groupings as seen in Appendix Tables 4 and 5, recommendations for revisions of errors that lead to false negatives generally aim towards a broader definition of asthma from a clinical perspective, while recommendations for revisions of errors that lead to false positives generally aim towards improving data capture and provenance.
From the perspective of a computable phenotype, the functional definition of asthma as commonly practiced in a healthcare setting may need to be expanded or adapted to accommodate for datapoints or features in forms more typically captured in an EHR. For instance, longitudinal use of certain asthma medications is a sufficient but not a necessary criterion of asthma - some asthma patients may not frequently use their medication for it to appear more than once on their charts. The particular demands of a computable phenotype that makes it different from a phenotype defined functionally is an area of ongoing interrogation. Also, there is inherent uncertainty surrounding the reliability of included features. For example, computable phenotypes that use ICD codes rather than specific asthma symptoms or labs rely on accurate physician diagnosis of the condition. Another example is the inaccuracy induced by the usage of a text-based feature, as seen in the PheKB algorithm.
Asthma in particular is challenging to model because of the complex diagnostic criteria that includes clinical symptoms, underlying inflammation and airflow limitation, showing phenotypic variation and overlap of other conditions. While pulmonary function testing is encouraged to determine airflow limitation, it is not routinely performed in practice.37 Accommodating population and institutional differences across populations is an important consideration when making design choices for computable phenotypes for asthma.
Limitations
Limitations to our study include that ambiguity does exist when determining a true asthma case, which may impact the reference standard to a degree. In addition, the final subset of patients we analyzed were what we determined to be unambiguous cases and were from a pre-existing cohort of patients with at least one ICD code related of asthma so the proportion of positive asthma cases in our final analysis set (~25%) is higher than what we would expect to see in the actual population. Also, the removal of ambiguous cases stemmed from one of the original goals of comparing the performance of the two algorithms. Measuring the performance on cases that even human reviewers found ambiguous would not give a meaningful metric as to whether the algorithms performed “correctly” on those cases. However, the study of such ambiguous cases in the context of automated phenotyping algorithms is of interest and will be examined in our future work. The mapping of ICD-9 to ICD-10 codes may also have affected our results. We did not have duplicate review of all cases and reviewers were not blinded to the results of the computable phenotypes, although they were instructed to not take it into consideration and provide an explanation for their label determination. Computable phenotypes are inherently limited by the availability of data within the system and human design. False positive identification of asthma cases occurs if all exclusion diagnoses are not taken into consideration. The performance of both computable phenotypes also faltered due to inability to incorporate detailed free-text. Use of text as a phenotype feature would require more powerful natural language processing methodologies. In addition, computable phenotypes that use an ICD code as a rule will not be able to identify missed or incorrectly diagnosed cases.
CONCLUSIONS
To understand how a computable phenotype for asthma might be built, two computable phenotypes were validated via comprehensive chart review. Through clinicians’ reviews, we identified four major categories of errors for the CAPriCORN and PheKB algorithms. Design decisions in a standardized asthma computable phenotype will affect performance across different populations and clinical environments such that multiple optimal configurations exist depending on the functional goal of the computable phenotype and that trade-offs will need to be considered. There is room to improve existing computable phenotypes if such algorithms are to reach a level of performance acceptable to common clinical implementation. Future considerations to identify asthma cases in the EHR include natural language processing and predictive algorithms.
Supplementary Material
ACKNOWLEDGEMENTS:
Javier Sanz and the UCLA Clinical and Translational Science Institute (CTSI) Data Core; Dr. Sleiman, CHOP. The authors would like to acknowledge the UCLA CTSI (UL TR001881) and the Institute for Precision Health (IPH) for their support in this project.
Footnotes
CONFLICT OF INTEREST: The authors declare that they have no conflicts of interest in the research.
Contributor Information
Mindy K. Ross, Department of Pediatrics, University of California Los Angeles, Los Angeles, USA
Henry Zheng, Department of Radiological Sciences, University of California Los Angeles, Los Angeles, USA.
Bing Zhu, Department of Radiological Sciences, University of California Los Angeles, Los Angeles, USA.
Ailina Lao, University of California Los Angeles, Los Angeles, USA.
Hyejin Hong, University of California Los Angeles, Los Angeles, USA.
Alamelu Natesan, Department of Pediatrics, University of California Los Angeles, Los Angeles, USA.
Melina Radparvar, Department of Pediatrics, University of California Los Angeles, Los Angeles, USA.
Alex AT Bui, Department of Radiological Sciences, University of California Los Angeles, Los Angeles, USA.
REFERENCES
- 1.Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014;21(2):221–230. PMC3932460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shang N, Liu C, Rasmussen LV, et al. Making work visible for electronic phenotype implementation: Lessons learned from the eMERGE network. J Biomed Inform. 2019;99:103293. PMC6894517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mo H, Thompson WK, Rasmussen LV, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J Am Med Inform Assoc. 2015;22(6):1220–1230. PMC4639716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016;23(6):1046–1052. PMC5070514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.NCQA. National Centers for Quality Assurance. HEDIS and Performance Measurement. https://www.ncqa.org/hedis/. Accessed December 6, 2020
- 6.Centers for Medicare and Medicaid Services. Chronic Conditions Data Warehouse. http://www.ccwdata.org/index.htm. Accessed December 6, 2020
- 7.AHRQ. Agency for Healthcare Research and Quality. Healthcare Cost and Utilization Project [Google Scholar]
- 8.Thompson WK, Rasmussen LV, Pacheco JA, et al. An evaluation of the NQF Quality Data Model for representing Electronic Health Record driven phenotyping algorithms. AMIA Annu Symp Proc. 2012;2012:911–920. PMC3540514 [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang G, Kiefer RC, Rasmussen LV, et al. Developing a data element repository to support EHR-driven phenotype algorithm authoring and execution. J Biomed Inform. 2016;62:232–242. PMC5490836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pletcher MJ, Forrest CB, Carton TW. PCORnet’s Collaborative Research Groups. Patient Relat Outcome Meas. 2018;9:91–95. PMC5811180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.OHDSI. Observational Health Data Sciences and Informatics. OHDSI Gold Standard Phenotype Library Architecture. https://www.ohdsi.org/2019-us-symposium-showcase-105/. Accessed January 6, 2020
- 12.Pacheco JA, Avila PC, Thompson JA, et al. A highly specific algorithm for identifying asthma cases and controls for genome-wide association studies. AMIA Annu Symp Proc. 2009;2009:497–501. PMC2815460 [PMC free article] [PubMed] [Google Scholar]
- 13.Geva A, Gronsbell JL, Cai T, et al. A Computable Phenotype Improves Cohort Ascertainment in a Pediatric Pulmonary Hypertension Registry. J Pediatr. 2017;188:224–231 e225. PMC5572538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashyap R, Sarvottam K, Wilson GA, et al. Derivation and validation of a computable phenotype for acute decompensated heart failure in hospitalized patients. BMC Med Inform Decis Mak. 2020;20(1):85. PMC7206747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trivedi M, Denton E. Asthma in Children and Adults-What Are the Differences and What Can They Tell us About Asthma? Front Pediatr. 2019;7:256. PMC6603154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amado MC, Portnoy JM. Diagnosing asthma in young children. Curr Opin Allergy Clin Immunol. 2006;6(2):101–105 [DOI] [PubMed] [Google Scholar]
- 17.National Asthma E, Prevention P. Expert Panel Report 3 (EPR-3): Guidelines for the Diagnosis and Management of Asthma-Summary Report 2007. J Allergy Clin Immunol. 2007;120(5 Suppl):S94–138 [DOI] [PubMed] [Google Scholar]
- 18.Reddel HK, FitzGerald jM, Bateman ED, et al. GINA 2019: a fundamental change in asthma management: Treatment of asthma with short-acting bronchodilators alone is no longer recommended for adults and adolescents. Eur Respir J. 2019;53(6) [DOI] [PubMed] [Google Scholar]
- 19.Cabana MD, Slish KK, Nan B, Clark NM. Limits of the HEDIS criteria in determining asthma severity for children. Pediatrics. 2004;114(4):1049–1055 [DOI] [PubMed] [Google Scholar]
- 20.CDC. Centers for Disease Control. Most Recent National Asthma Data. 2017; https://www.cdc.gov/asthma/most_recent_national_asthma_data.htm
- 21.Nurmagambetov T, Kuwahara R, Garbe P. The Economic Burden of Asthma in the United States, 2008–2013. Ann Am Thorac Soc. 2018;15(3):348–356 [DOI] [PubMed] [Google Scholar]
- 22.Collaco JM, Abman SH. Evolving Challenges in Pediatric Pulmonary Medicine. New Opportunities to Reinvigorate the Field. Am J Respir Crit Care Med. 2018; 198(6):724–729 [DOI] [PubMed] [Google Scholar]
- 23.Afshar M, Press VG, Robison RG, et al. A computable phenotype for asthma case identification in adult and pediatric patients: External validation in the Chicago Area Patient-Outcomes Research Network (CAPriCORN). J Asthma. 2017:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almoguera B, Vazquez L, Mentch F, et al. Identification of Four Novel Loci in Asthma in European American and African American Populations. Am J Respir Crit Care Med. 2017;195(4):456–463. PMC5378422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medicare Cf, Services M, Statistics NCfH. ICD-10-CM official guidelines for coding and reporting FY 2018. 2019 [Google Scholar]
- 26.“Global strategy for asthma management and prevention: GINA executive summary.” Bateman ED, Hurd SS, Barnes PJ, Bousquet J, Drazen JM, FitzGerald JM, Gibson P, Ohta K, O’Byrne P, Pedersen SE, Pizzichini E, Sullivan SD, Wenzel SE and Zar HJ Eur Respir J 2008; 31: 143–178. Eur Respir J. 2018;51(2) [DOI] [PubMed] [Google Scholar]
- 27.Xi N, Wallace R, Agarwal G, et al. Identifying patients with asthma in primary care electronic medical record systems Chart analysis-based electronic algorithm validation study. Can Fam Physician. 2015;61(10):e474–483. PMC4607352 [PMC free article] [PubMed] [Google Scholar]
- 28.Engelkes M, Afzal Z, Janssens H, et al. Automated identification of asthma patients within an electronical medical record database using machine learning. European Respiratory Journal. 2012;40 [Google Scholar]
- 29.Afzal Z, Engelkes M, Verhamme KMC, et al. Automatic generation of case- detection algorithms to identify children with asthma from large electronic health record databases. Pharmacoepidem Dr S. 2013;22(8):826–833 [DOI] [PubMed] [Google Scholar]
- 30.Dexheimer JW, Abramo TJ, Arnold DH, et al. Implementation and evaluation of an integrated computerized asthma management system in a pediatric emergency department: a randomized clinical trial. Int J Med Inform. 2014;83(11):805–813. PMC5460074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu ST, Sohn S, Ravikumar KE, et al. Automated chart review for asthma cohort identification using natural language processing: an exploratory study. Ann Allergy Asthma Immunol. 2013;111(5):364–369. PMC3839107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozyrskyj AL, HayGlass KT, Sandford AJ, et al. A novel study design to investigate the early-life origins of asthma in children (SAGE study). Allergy. 2009;64(8):1185–1193 [DOI] [PubMed] [Google Scholar]
- 33.Vollmer WM, O’Connor EA, Heumann M, et al. Searching multiple clinical information systems for longer time periods found more prevalent cases of asthma. J Clin Epidemiol. 2004;57(4):392–397 [DOI] [PubMed] [Google Scholar]
- 34.Donahue JG, Weiss ST, Goetsch MA, et al. Assessment of asthma using automated and full-text medical records. J Asthma. 1997;34(4):273–281 [DOI] [PubMed] [Google Scholar]
- 35.Premaratne UN, Marks GB, Austin EJ, Burney PG. A reliable method to retrieve accident & emergency data stored on a free-text basis. Respir Med. 1997;91(2):61–66 [DOI] [PubMed] [Google Scholar]
- 36.Richesson RL, Sun J, Pathak J, Kho AN, Denny JC. Clinical phenotyping in selected national networks: demonstrating the need for high-throughput, portable, and computational methods. Artif Intell Med. 2016;71:57–61. PMC5480212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sd Aaron, Boulet LP Reddel HK, Gershon AS. Underdiagnosis and Overdiagnosis of Asthma. Am J Respir Crit Care Med. 2018;198(8):1012–1020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.