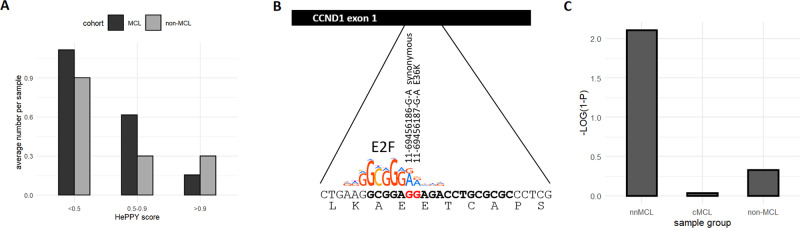
Fig. 6. Prevalence of synonymous and benign mutations in the first exon of CCND1 in MCL.
A The number of samples with mutations in exon1 of CCND1 by HePPY score is shown. HePPY is a predictor of missense variant pathogenicity (10.1182/blood-2019-128488). Low HePPY scores indicate benign amino acid changes. Deleterious mutations are associated with HePPY scores close to 1. B The sequence coding for CCND1 L32 to S41 is shown. Two frequently observed mutations in the first exon of CCND1 are overlapping an E2F-binding site. The red guanine bases are often changed to adenine, which favors E2F binding. C Significance of the proportion of synonymous mutations in the sample groups displayed on the x-axis. P values were calculated using the cumulative hypergeometric distribution. The negative decadic logarithm of (1−P) is shown. Values >2 are considered significant.