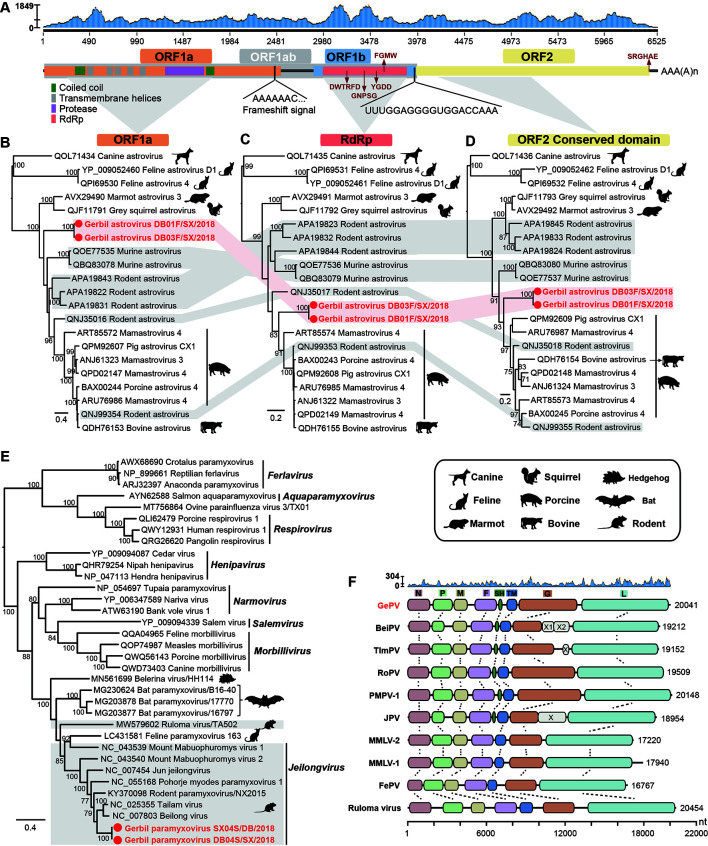
Figure 1.
Genome characterization and evolutionary relationships of GeAstV and GePV
A: Schematic representation and read abundance of the genome of GeAstV. The characteristic amino acid motifs are highlighted with red arrows. B–D: Maximum likelihood phylogenies of GeAstV and representative members of the genus Mamastrovirus were inferred based on the amino acid alignments of ORF1a (B), RdRp (C), and ORF2 (D). The two gerbil astrovirus sequences are highlighted in red with red solid circles. The grey-shaded regions show the sequences with phylogenetic incongruence and the rodent associated astrovirus. E: Maximum likelihood phylogeny of the L protein of gerbil paramyxovirus and the representative members of the Paramyxoviridae, including those from the genera Ferlavirus, Aquaparamyxovirus, Respirovirus, Henipavirus, Narmovirus, Salemvirus, Morbillivirus, and Jeilongvirus. The rodent associated viruses are highlighted with a grey background. F: The genome of GePV and nine paramyxoviruses from the genus Jeilongvirus: Beilong virus (BeiPV), Tailam virus (TlmPV), Rodent paramyxovirus (RoPV), Pohorje myodes paramyxovirus 1 (PMPV-1), Jun jeilongvirus (JPV), Mount Mabu Lophuromys virus 1 (MMLV-1) and 2 (MMLV-2), Feline paramyxovirus(FePV), and Ruloma virus. Read abundance of the genome of GePV are shown on the top, followed by open reading frames (ORFs) colored according to their putative functions. The numbers to the right of each virus indicate the length of the virus genome. The genomes are drawn to a unified length scale shown at the bottom. Homologous gene regions are connected by dotted lines. B–E: The trees were midpoint rooted for clarity. Bootstrap values of >70% are shown for key nodes.