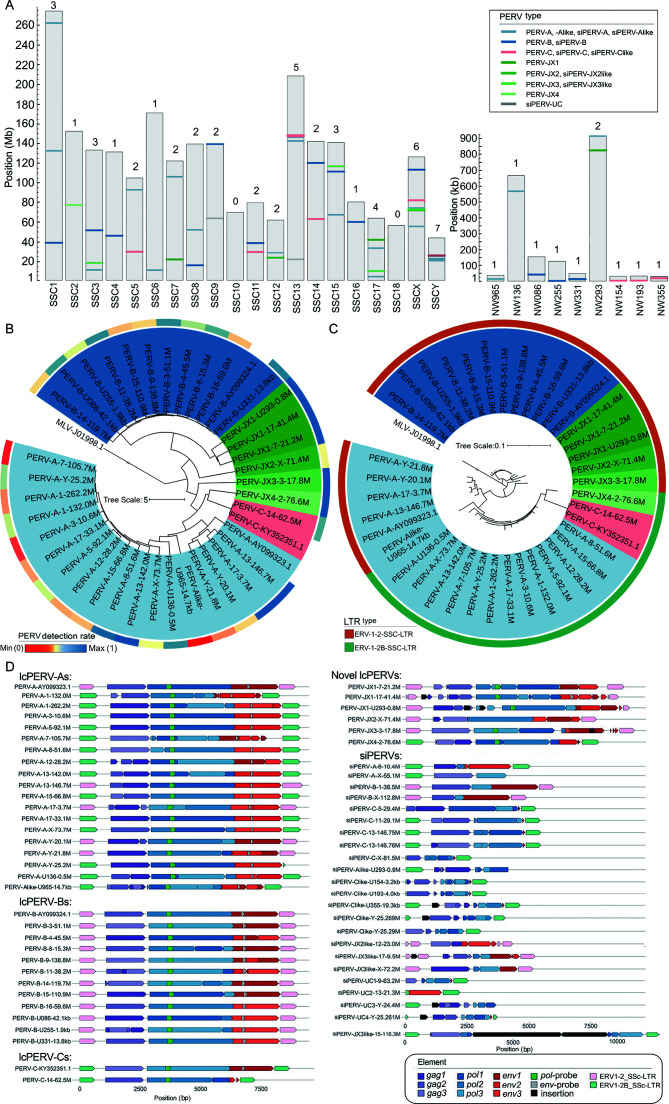
Figure 2.
59 PERVs in the S. Scrofa reference genome
A: The positions of PERVs in the reference genome. NW965: NW_018084965; NW136: NW_018085136; NW086: NW_018085086; NW255: NW_018085255; NW331: NW_018085331; NW293: NW_018085293; NW154: NW_018085154; NW193: NW_018085193; NW355: NW_018085355. siPERV: short incomplete PERV. B: Maximum-likelihood (ML) phylogenetic tree of 36 long complete or near-complete PERVs (lcPERVs) without LTR. In the tree, 3 complete PERVs (PERV-A: AY099323.1, PERV-B: AY099324.1, and PERV-C: KY352351.1) were included, and an MLV (MLV-J01998.1) was set as the outgroup. The color on the outer circle represents the detection rate of PERVs on the genome by 10 breeds (n=63). The bluer the color, the more pigs have this PERV copy, and the redder the color, the fewer pigs. C: Maximum-likelihood (ML) phylogenetic tree of 36 lcPERVs with LTRs. The color on the outer circle represents LTR types. D: ORF prediction of 62 PERVs and their LTRs. 59 PERVs were identified from the swine reference genome Build 11.1, and 3 complete PERVs (PERV-A: AY099323.1, PERV-B: AY099324.1, and PERV-C: KY352351.1) were downloaded from the NCBI Genbank database. ORFs of gag, pol, env were predicted in three forward reading frames. For example, gag1 means gag ORF predicted in forward frame 1; gag2 means gag ORF predicted in forward frame 2; gag3 means gag ORF predicted in forward frame 3. The sequences of pol-probe and env-probe were downloaded from the reference (Yang et al., 2015).