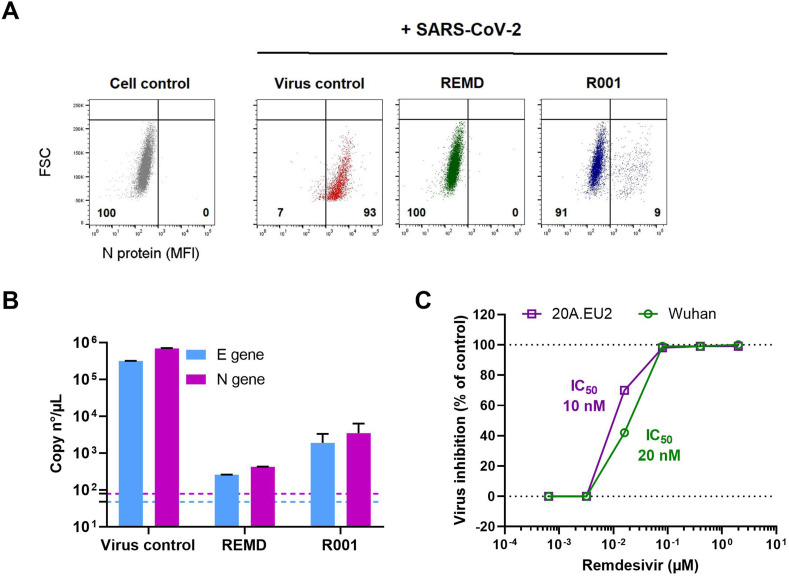
Fig. 5.
Analysis of antivirals in SARS-CoV-2 infected U87.ACE2+ cells by flow cytometry and RT-qPCR. A) U87.ACE2+ cells were exposed to SARS-CoV-2 (Wuhan strain, MOI 1) in absence (Virus control) or presence of inhibitors (2 μM Remdesivir or 10 μg/ml antibody R001). At 2 d p.i. cells were collected and stained for intracellular nucleoprotein (N) expression. Dot plots show N expression in noninfected (lower left quadrant) and infected (lower right quadrant) cells, based on the analysis of 5,000–6,000 cells by flow cytometry. The numbers in each quadrant refer to the distribution of the cells (i.e., percentage of total cell population). The dot plot in grey (left panel) represents the noninfected cell control. B) U87.ACE2+ cells were exposed to SARS-CoV-2 (Wuhan strain, MOI 0.3) for 2 h in absence (Virus control) or presence of inhibitors (2 μM Remdesivir or 10 μg/ml antibody R001). Supernatant (= virus input) was then removed, and cells were washed in PBS and given fresh medium. In the conditions with the antiviral treatment, compound was administered together with the virus, and administered again after the wash step. At day 2 p.i., supernatant was collected and RT-qPCR was performed to quantify the copy numbers of the N gene and the E gene. The dotted colored lines refer to the residual amount (background) of virus that attached nonspecifically to the cells and was measured immediately after the wash step (2 h p.i.). For each sample, the average (mean ± SD) of two technical replicates is given (n = 2), plotted on a log10 scale. C) Same as in (B) but for a dilution range of remdesivir. U87.ACE2+ cells were exposed to two different variants of concern of SARS-CoV-2 (MOI 0.3), as indicated. RT-qPCR data of the N gene were used to calculate the % inhibition of viral replication and to plot a concentration-response curve for Remdesivir. The calculated IC50 value of Remdesivir for each virus strain is indicated in the matching color.