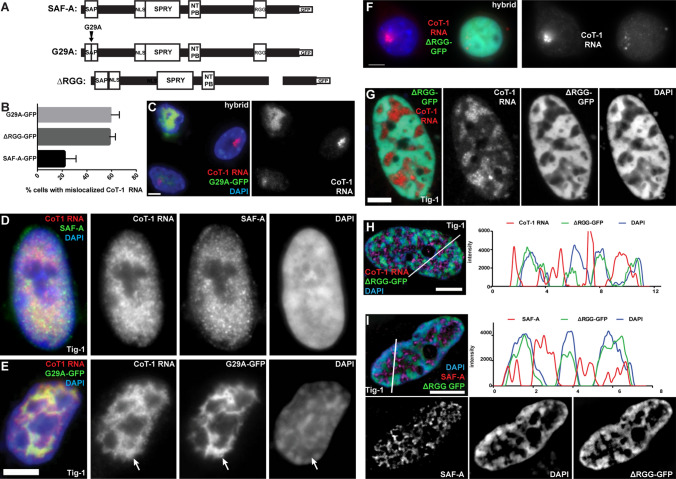
Fig. 3.
Both SAF-A DNA binding and RGG binding domain mutants displace C0T-1 RNA and alters DNA morphology. For all images: color channels are separated in black and white. Scale bars 5 μm. Cell types: mouse/human hybrid cells with human chromosome 4 (Hybrid) & normal human fibroblasts (Tig-1). A Map of full-length SAF-A, and the G29A and ∆RGG mutants. B Both SAF-A mutants affect C0T-1 RNA localization to chromatin (G29A: P = 0.001, RGG: P = 0.004). Error bars, standard deviation of the mean. Wildtype SAF-A-GFP also affected C0T-1 RNA localization when it was grossly overexpressed (in ~ 20% of cells). C Expression of the G29A mutant releases C0T-1 RNA from the human chromosome in hybrid cells, while non-transfected neighboring cells are unaffected. D–E Normal C0T-1 RNA distribution in fibroblasts (D) is altered when the G29A mutant is expressed (E). C0T-1 RNA localizes with G29A-GFP and not with DAPI DNA staining (arrows), and DNA condensation is altered. F Expression of the ∆RGG mutant mis-localizes human C0T-1 RNA from the human chromosome in hybrid cells compared to a neighboring cell that was not transfected. G C0T-1 RNA is released from chromatin in fibroblasts expressing the ∆RGG SAF-A mutant, and DNA compaction is altered. H SIM image and linescan showing relative distributions and intensity of DAPI DNA, C0T-1 RNA, and ∆RGG SAF-A mutant in a fibroblast nucleus. I SIM image and linescan showing distribution of DAPI DNA, endogenous SAF-A and ∆RGG SAF-A mutant in a fibroblast nucleus