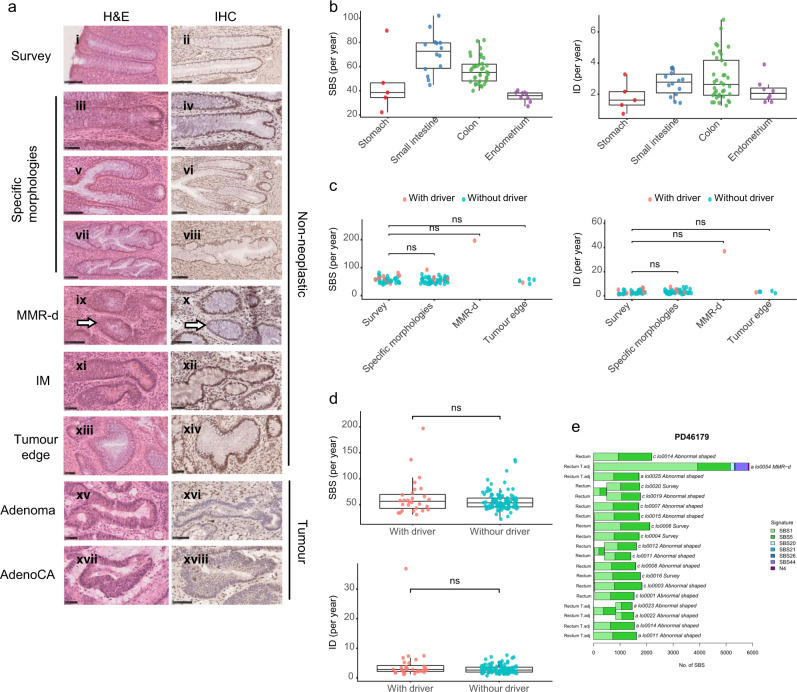
Fig. 1. Mutation burden in the non-neoplastic crypts in LS patients.
a Representative images of H&E and IHC staining of crypts with different morphologies. Except for the MMR deficient crypt (indicated by white arrows), images of H&E and IHC did not represent the same crypt. MMR protein antibodies were used according to the germline MMR gene mutation in respective patients. MMR-d: MMR-deficient; IM: intestinal metaplasia; AdenoCA: adenocarcinoma. Scale bar: 100 μm (i, ii, v, vi, vii, viii, ix, x, xvi, xviii); 50 μm (iii, iv, xi, xii, xiii, xiv, xv, xvii). b–d Boxplots showing the 1.5X interquartile range (whiskers), the first and third quartile (bounds of box), and the median (centre)of the data. Each data point represented a crypt. Two-sided Wilcoxon test: ns = non-significant. b Single base substitutions (SBS) and insertions and deletions (ID) burden of survey crypts from the stomach (n = 5, red), the small intestine (n = 14, blue), the colon (n = 37, green) and the endometrium (n = 8, purple). c SBS and ID burden in different types of colonic crypts (Survey (n = 37), Specific morphologies (n = 50), MMR-d (n = 1), Tumour edge (n = 5)). Crypts with and without driver mutations are indicated in pink and blue, respectively. d SBS and ID burden in non-neoplastic crypts with (n = 28, pink) and without (n = 104, blue) cancer driver mutations. e A phylogenetic tree showing the clonal relationship between non-neoplastic crypts in PD46179. Branch lengths correspond to the number of SBS mutations. SBS mutational signatures are mapped onto tree branches. Each crypt is annotated with its identifier, crypt type, and tissue origin.