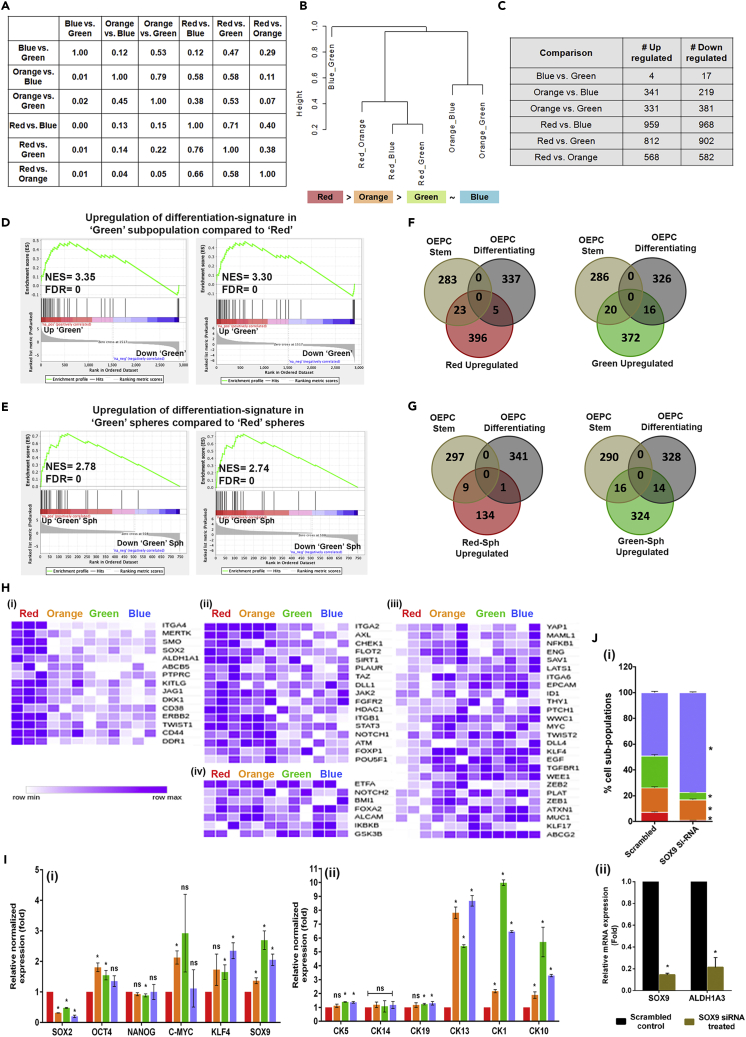
Figure 4.
Subpopulation specific transcriptome analysis
(A and B) Overlap in DE-Gs of different pairwise comparisons. Proportion of overlap between each pairwise comparison (A).Hierarchical structure of different cell subpopulations (B).
(C) Pairwise comparisons of differentially expressed genes of four subpopulations of GBC02 cell line.
(D) Gene set enrichment analysis (GSEA) with genes up-regulated in ‘Green’ subpopulation as compared to ‘Red’ subpopulation from GBC02 monolayer cultures showing enrichment of genes involved in Keratinization and Cornified envelope formation.
(E) Gene set enrichment analysis (GSEA) with genes up-regulated in Green subpopulation as compared to Red subpopulation from GBC02 3D spheroids showing enrichment of genes involved in keratinization and cornified envelope formation.
(F and G) Venn diagram showing overlap of basal OEPC and basal differentiating gene sets with Red versus Green comparison upregulated genes in Red and Green sorted subpopulations monolayer cultures (F) and Red and Green 3D spheroids from GBC02 cell line (G).
(H) Heatmaps of mean normalized expression values from GBC02 monolayer cultures for genes expressed distinctly in (1) ‘Red’ (2) both ‘Red’ and ‘Orange’ subpopulations (3) commonly in ‘Orange’, ‘Green’ and ‘Blue’ subpopulations and (4) among all the four subpopulations.
(I) qRT-PCR of the four sorted subpopulations from GBC02 monolayer cultures for various (1) stemness genes and (2) differentiating cytokeratins.
(J) (1) Graph showing decreased frequency in ALDHHigh subpopulations in SCC-070 cell line upon siRNA mediated silencing of SOX9. Statistical comparisons were made between each subpopulation in scrambled versus SOX9 siRNA treatment conditions (ex; Green scrambled versus Green SOX9 siRNA). Error bars represent mean ± SEM from three biological repeats and ∗ indicates p value ≤0.05. (2) qRT-PCR after siRNA knockdown of SOX9 in SCC-070 cell line. Error bars represent mean ± SD from three biological repeats and ∗ indicates p value ≤0.05 for qRT-PCR experiments.