Figure 5.
The computational model quantitatively replicates the experiments
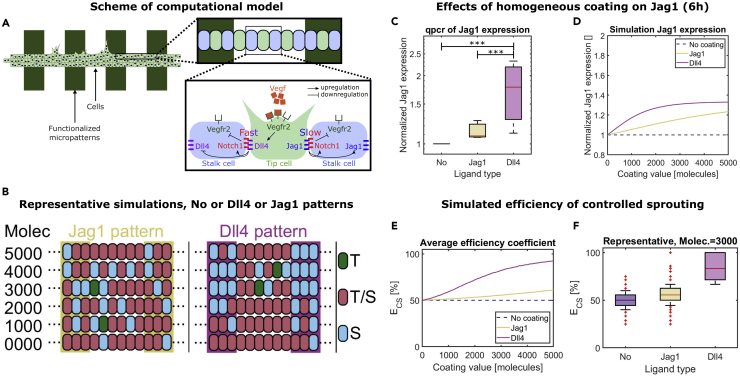
(A) Due to periodicity, cell-cell signaling was modeled only for a small portion of the cells on the patterned substrate. The first enlargement shows the cells that were analyzed in the simulations. The second enlargement shows the main features of the simulated signaling network.
(B) Representative computational results, with each row corresponding to one simulation, with different ligand concentrations on the micropatterns (decreasing from top to bottom). Without ligands on the patterns (bottom row) most cells have a tip/stalk (T/S) hybrid phenotype (red). With increasing concentration, Dll4 induces the stalk (S, blue) phenotype for cells on top of the lines, whereas Jag1 does not have evident effects. Only few tip (T, green) cells were observed.
(C and D) The effects of homogeneous coating of Fc, Jag1, or Dll4 on the gene expression of Jag1 were compared between experimental (C) and computational (D) data for the time period of 6 h. qPCR revealed a significant difference of Jag1 expression for Dll4 versus Fc samples (p < 0.0001) and for Dll4 versus Jag1 samples (p < 0.001). For the computational data, averages over 10,000 simulations for each concentration are reported.
(E) The efficiency of controlled sprouting was computed for simulations of cells on patterned lines with varying ligand densities. The graph reports the averages of the computed values over 10,000 simulations.
(F) Boxplots obtained for the simulations with Dline = Jline = 3000 molecules. The simulations captured the higher spatial control of Dll4 lines, compared with Jag1 lines, on the location of the sprouts (E and F). In (C) and (F), data are represented as boxplots, where the boxes span from the 25th until the 75th percentile, the whiskers extend down to the 10th percentile and up to the 90th percentile, and the horizontal lines represent the median values.