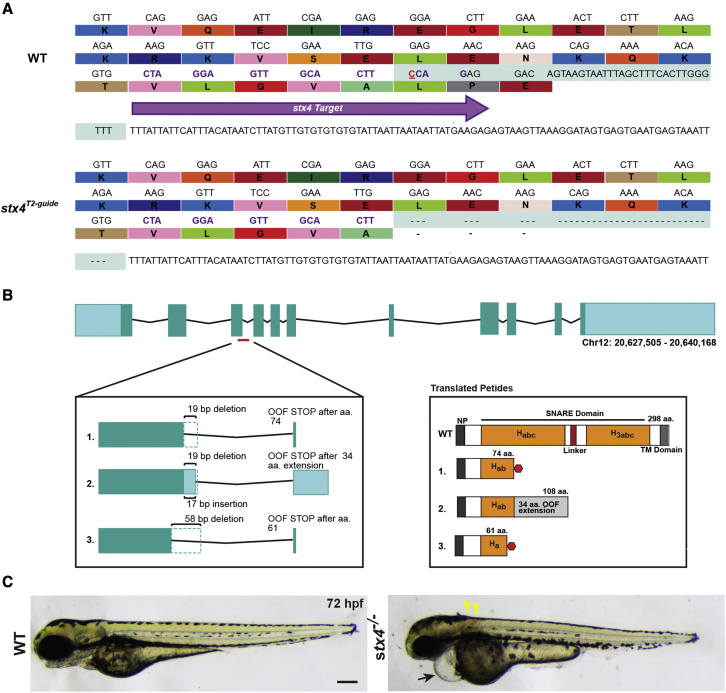
Figure 2.
Generation of stx4 mutant zebrafish
(A) CRISPR-Cas9 was used to generate a zebrafish stx4 loss-of-function allele that creates a 38-bp deletion (red underlined cytosine marks the endonuclease cut site), eliminating the splice donor site of exon 3 (protospacer denoted by purple arrow and codons; the last three codons at the splice junction are eliminated in stx4 mutants).
(B) Schematic of transcripts showing the three alternative splice variants generated from this allele. All three transcripts are predicted to be out-of-frame (OOF; left) and to produce truncation products (translated peptides; right). Habc, antiparallel three-helix bundle stabilization domain; H3abc: coiled-coil SNARE homology domain; NP, N-terminal peptide, TM, transmembrane. Red polygon represents a stop codon in the 298 amino acid sized zebrafish Stx4 protein, resulting in early truncation products.
(C) Bright field images of 72 hpf WT/stx4+/− sibling and stx4 mutant larvae. Images of WT sibling embryos in all figures are stx4+/− sibling embryos. Black arrow indicates pericardial edema and linear heart. Yellow arrowheads indicate hemorrhages. stx4 heterozygotes are overtly indistinguishable from WT. Scale bar, 200 μm.