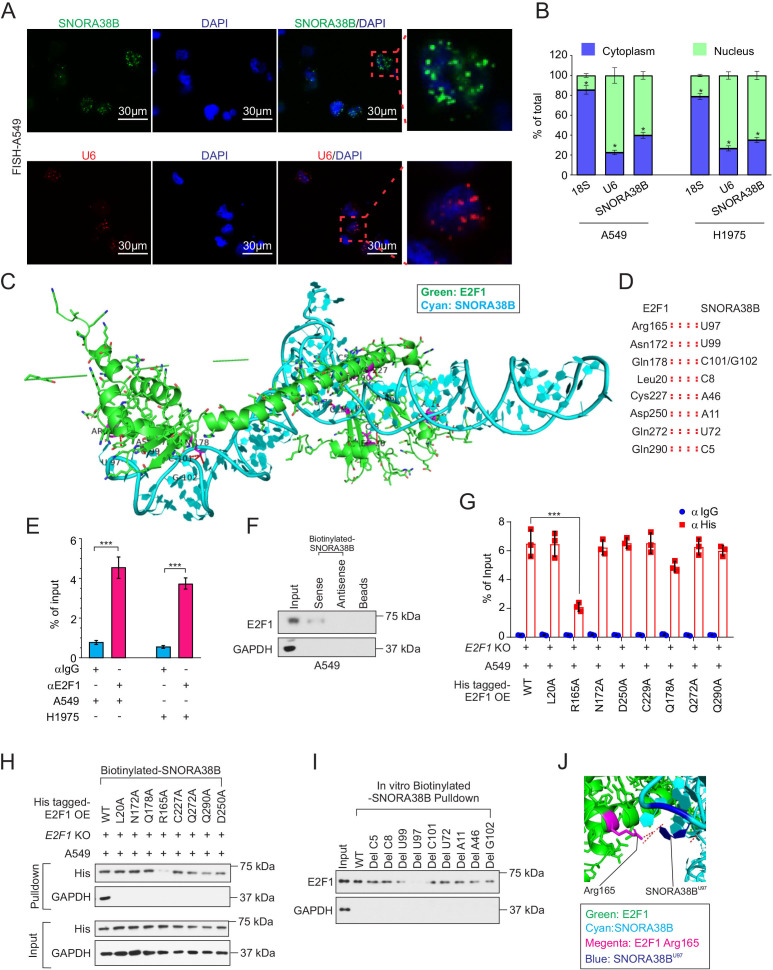
Figure 6.
SNORA38B directly bound with E2F1. (A) Position of SNORA38B/U6 in A549 cells shown by RNA fluorescence in situ hybridization analysis (FISH). Bar=30 µm. (B) Position of SNORA38B in A549 and H1975 cells shown by nuclear and cytoplasmic RNA fractionation analysis. 18S and U6 were used as cytoplasmic and nuclear internal controls, respectively. (C–D) The whole view of SNORA38B’s interaction with the regulatory domain of E2F1. Green: E2F1; Cyan: SNORA38B. (E) RIP detecting the binding enrichment of E2F1 with SNORA38B in A549 and H1975 cells; anti-IgG as controls. (F) RNA pulldown assay detected the interaction of SNORA38B and E2F1 in A549 cells using antisense small nucleolar RNA and beads as controls. (G) RIP assay detecting the binding activities between his-tagged E2F1-WT or E2F1-mutants with SNORA38B in E2F1-WT or E2F1-mutants treated E2F1-KO A549 cells. (H) RNA pulldown assay detected the interaction of SNORA38B and E2F1-WT or E2F1-mutants in E2F1 WT or mutants treated E2F1-KO A549 cells. (I) In vitro RNA pulldown assay detected the interaction of SNORA38B-WT or SNORA38B-mutants; Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a negative control. (J) Magnified view of SNORA38B’s interaction with the regulatory domain of E2F1. SNORA38B U97 forms hydrogen bonds and polar interaction with Arg165. Color representation: green indicates E2F1; cyan indicates SNORA38B; magenta indicates Arg165; blue indicates SNORA38B U97. *p<0.05, ***p<0.001, means±SD was shown. Statistical analysis was subjected to Student’s t-test and one-way analysis of variance . Assays were conducted in triple. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; KO, knockout; OE, overexpress; RIP, RNA immunoprecipitation; SNORNA, small nucleolar RNA; WT, wild-type.