Extended Data Fig. 5 |. BSL1 Interferes the Interaction of BIN2-BASL in vitro.
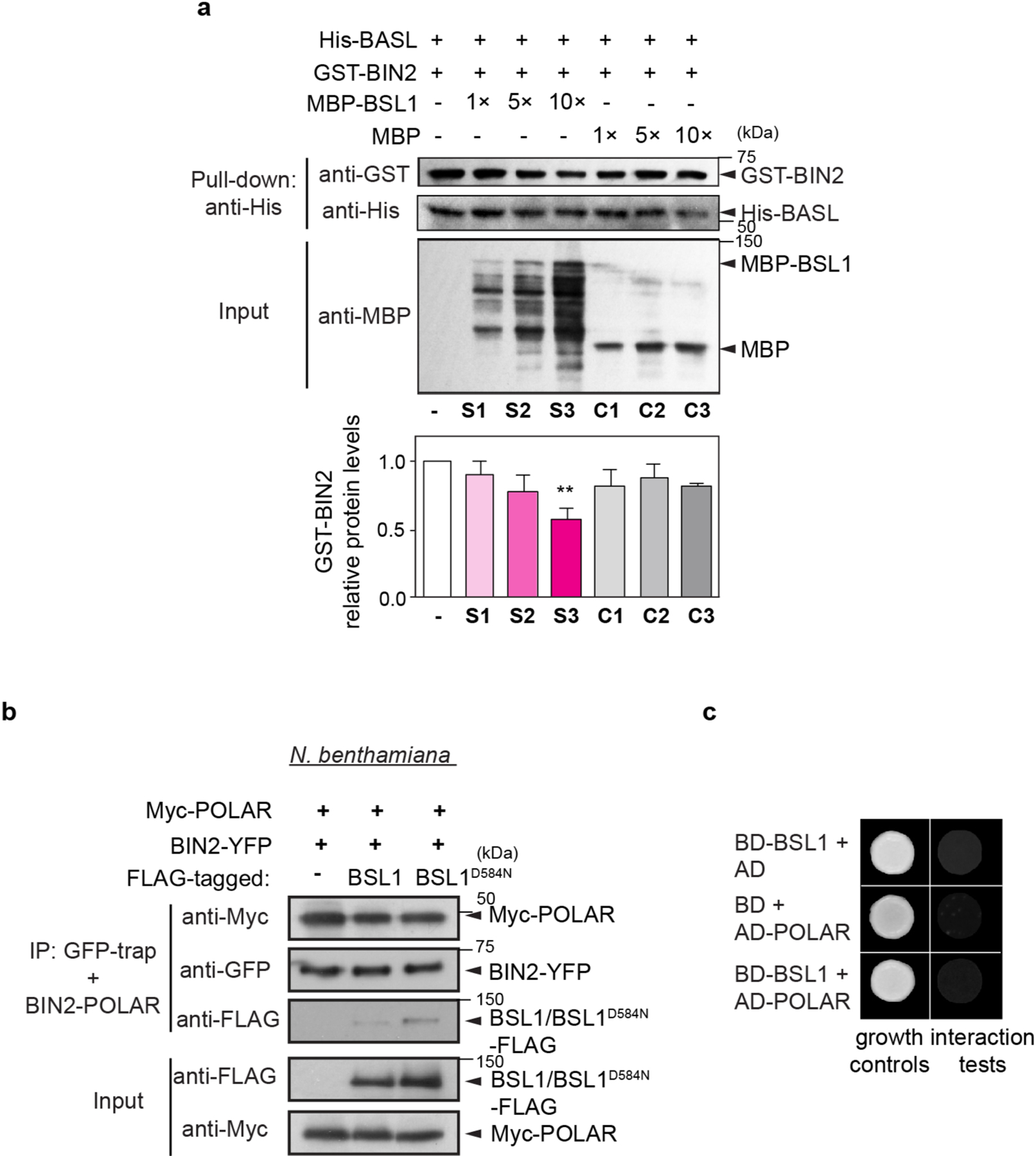
a, In vitro pull-down assays using recombinant proteins to test the BIN2-BASL interaction in the presence of an increasing amount of MBP-BSL1. His-BASL was used as bait and the amount of GST-BIN2 being pulled down reflects the interaction strength of BIN2-BASL. MBP was used as negative control. Histograms (below) show quantification of relative protein levels of BIN2 in the assay above. Results suggest the addition of MBP-BSL1 reduced the amount of BIN2 that interacted with BASL. Data are presented as mean ± SD. n = three independent experiments. Two-tailed Student’s t-tests. ** P < 0.005. b, Co-IP assays using purified fusion proteins produced by N. benthamiana leaves show physical association of BIN2-YFP with Myc-POLAR was not influenced by the presence of FLAG-tagged BSL1 or BSL1D584N. BIN2-YFP was used as bait. Immunoprecipitated proteins were detected by anti-Myc. Data represent results of experiments repeated three times. c, Results of yeast two-hybrid assays show no interactions between BSL1 and POLAR were identified. BD indicates Gal4 DNA-binding domain. AD indicates Gal4 activation domain. ‘Interaction tests’, assays performed using synthetic dropout medium (-Leu-Trp-His; 1 mM 3-AT added to suppress auto-activation); ‘growth controls’, assays performed using rich media (-Leu-Trp).
