Figure 1. Predicted coiled-coil propensity for the cardiac myosin rod.
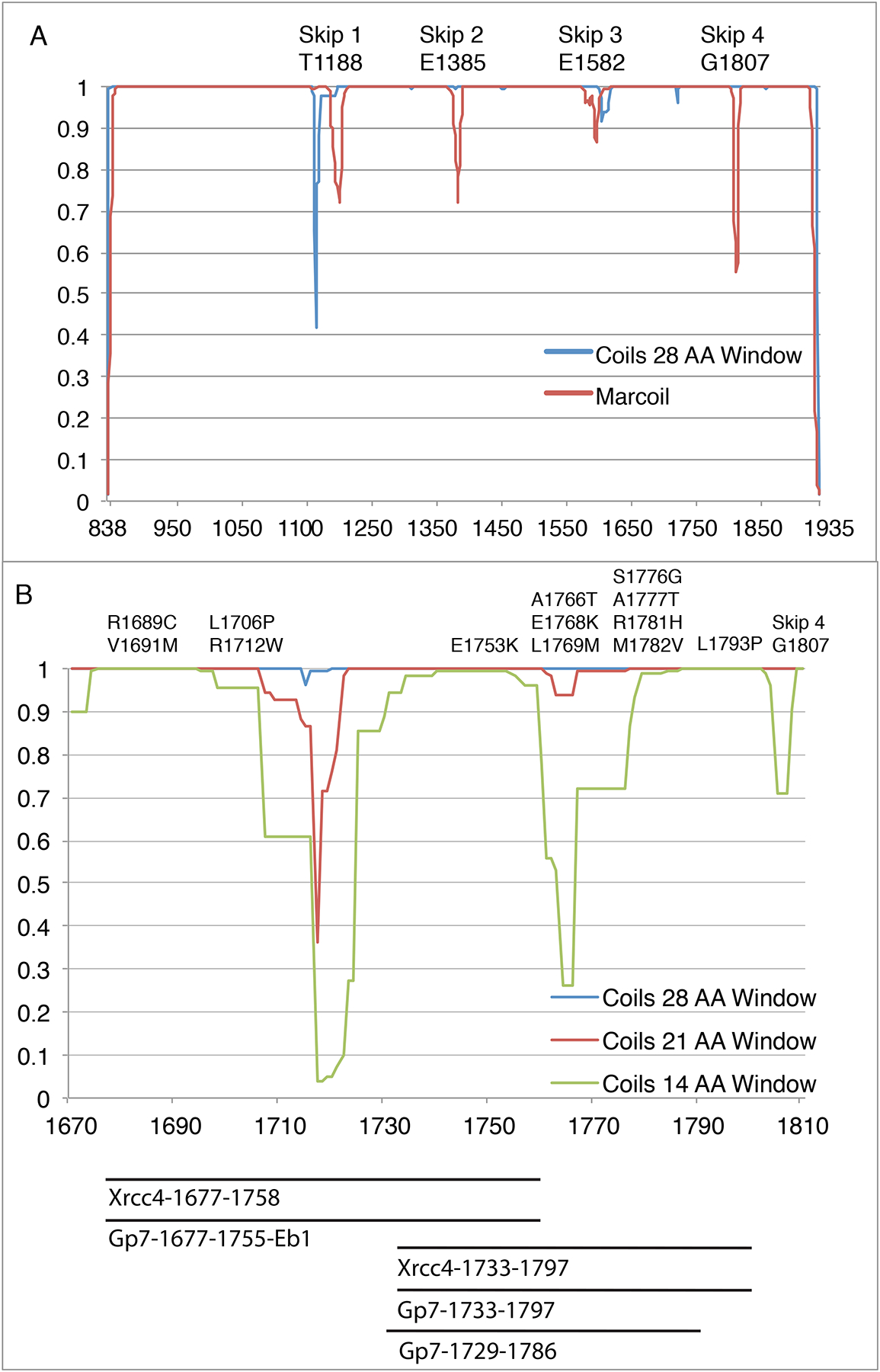
(A) Myosin rod coiled-coil prediction using a 28-amino acid window in COILS (blue) and Marcoil (Red). The COILS algorithm compares a sequence to a database of known parallel two-stranded coiled-coils and derives a similarity score (Lupas, 1996a; Lupas et al., 1991). The 28-amino acid window for coils smooths out most of the local variations in coiled coil propensity except for around the Skip residues which introduce a discontinuity. The Marcoil algorithm is based on a window-less Hidden Markov Model (Delorenzi and Speed, 2002). The propensities relate to the probability that a group of amino acids will adopt the structure of a coiled-coil, but do not indicate stability or structure of the resultant oligomeric state. (B) COILS prediction from myosin rod amino acids 1680–1810. This region contains 13 pathogenic mutation sites, as well as regions of poorly predicted coiled-coil.
