Fig. 1.
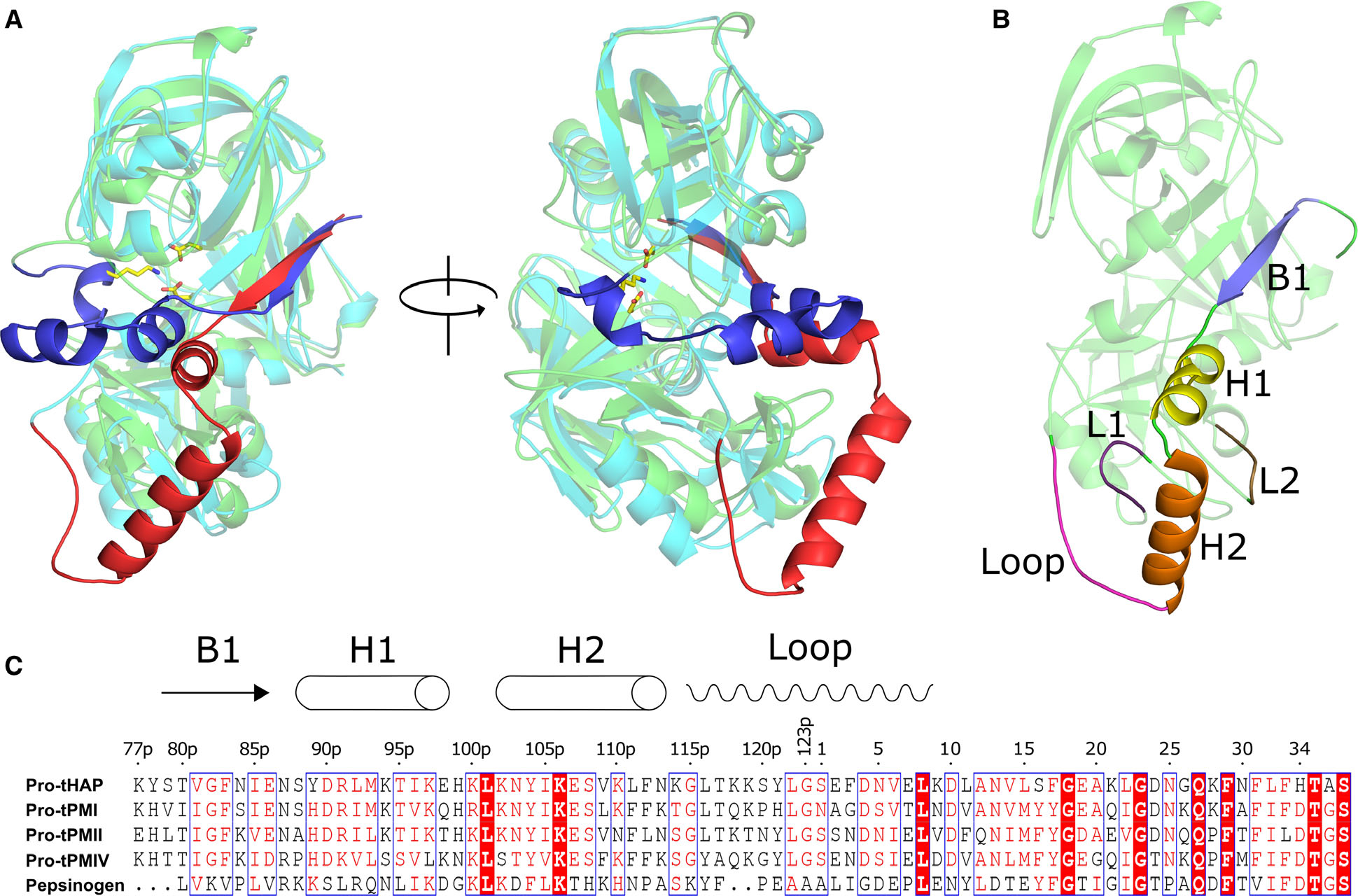
Structural and sequence alignment of the pro-tPMs and pepsinogen. (A) Structural superposition of Pv-pro-tPM (1MIQ) and porcine pepsinogen (3PSG), with their mature parts in cyan and green; truncated prosegments in red and blue, respectively. (B) Domain architecture of pro-tPMs; the mature polypeptide is colored green and the secondary structural elements of the truncated prosegment are distinctly colored. The protein structural representations have been generated with the PyMOL molecular graphics system (Schrödinger LLC, New York, NY, USA; version 2.3.2). (C) Multiple sequence alignment of pro-tPMs (uniprot accession number: pro-tHAP-Q9Y006; pro-tPMI-P39898; pro-tPMII-P46925; pro-tPMIV-W7FF86; porcine pepsinogen-P00791) and pepsinogen was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/), with the secondary structural elements of pro-tHAP shown on the top of the alignment. The highly conserved residues are shown as white on red background as generated by ESPript 3 [60]. The numbers on the top of the alignment represent the residue position in the pro-tHAP structure. The letter ‘p’ in the sequence numbering implies the residues in the prosegment.
