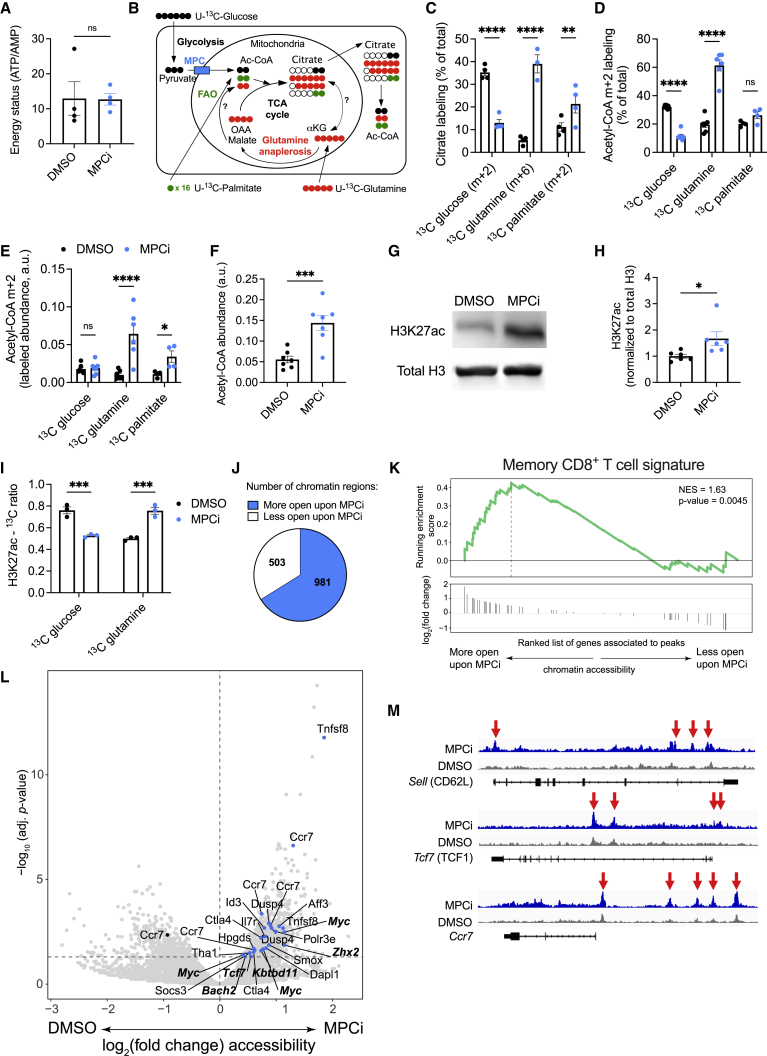
Figure 2.
MPC inhibition in CD8+ T cells induces alternative metabolic fluxes that results in an epigenetic crosstalk and memory imprinting
(A) Energy status (ATP/AMP), 72 h post-activation, based on mass spectrometry data (n = 4 biological replicates).
(B) Schematic representation of metabolite labeling patterns (OAA, oxaloacetate; αKG, α-ketoglutarate).
(C and D) Percentage of the indicated citrate isotope out of total citrate (C) or of m+2 acetyl-CoA out of total acetyl-CoA (D) using different heavy labeled substrates.
(E) Abundance of m+2 acetyl-CoA, derived from different heavy labeled substrates.
(F) Abundance of total cellular acetyl-CoA.
n = 4 biological replicates in (C) and n = 7 biological replicates in (D)–(F); pooled from 2 independent experiments.
(G and H) Representative western blot (G) and quantification shown as fold change compared with DMSO (H) (n = 6 biological replicates; pooled from 4 independent experiments).
(I) Incorporation of carbons derived from glucose or glutamine into the acetyl group on H3K27. Shown here is the ratio of the intensity of the 3rd isotopic peak (containing 2∗13C) over the monoisotopic peak (only 12C) of acetylated peptide KSAPATGGVKKPHR (see also Figures S2H and S2I) (n = 3 biological replicates).
(J) Number of chromatin regions associated with more open or closed regions upon MPCi treatment (n = 3 [DMSO] or 2 [UK5099] biological replicates).
(K) Gene set enrichment analysis of an MPEC signature (Dominguez et al., 2015).
(L) Volcano plot showing the genes associated to more closed (left) or more open (right) chromatin regions upon MPCi treatment. Genes associated with the MPEC signature in (K) are highlighted in blue, with genes previously associated to H3K27ac in bold (Gray et al., 2017).
(M) Representative ATAC-seq traces in and around the gene loci of Sell, Tcf7, and Ccr7. Red arrows highlight increased chromatin accessibility.
Data are represented as mean ± SEM. Statistics are based on unpaired, two-tailed Student’s t test (A, F, and H) or two-way ANOVA (C–E and I), ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, and ns (p > 0.05). See also Figures S1 and S2.