ABSTRACT
Candida auris is an emerging yeast species that has the unique characteristics of patient skin colonization and rapid transmission within health care facilities and the ability to rapidly develop antifungal resistance. When C. auris first started to appear in clinical microbiology laboratories, it could be identified only by using DNA sequencing. In the decade since its first identification outside of Japan, there have been many improvements in the detection of C. auris. These include the expansion of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) databases to include C. auris, the development of both laboratory-developed tests and commercially available kits for its detection, and special CHROMagar for identification from laboratory specimens. Here we discuss the current tools and resources that are available for C. auris identification and detection.
KEYWORDS: Candida auris, CHROMagar, antifungal susceptibility testing, fungal, testing
INTRODUCTION
Candida auris is a relatively new species of yeast, first identified in 2009, that has quickly spread across the world (1, 2). There are three features that make C. auris unique from other species of Candida: (i) antifungal resistance is the norm for C. auris rather than the exception (3–5), (ii) rather than primarily colonizing the gut, C. auris colonizes the skin, anterior nares, and other body sites of asymptomatic carriers, and (iii) C. auris is transmitted easily between patients in health care settings. Some patients can be asymptomatically colonized with C. auris for long periods of time, and these colonized patients contribute to environmental contamination and transmission within health care settings. The transmissibility of C. auris and the alarming statistic that 5 to 10% of colonized patients subsequently develop bloodstream infections make C. auris a serious public health problem (6). To complicate matters further, an overwhelming percentage of isolates, up to 99% in some jurisdictions, are resistant to at least one commonly used antimicrobial, and isolates have been identified that are resistant to azoles, echinocandins, and amphotericin B, severely limiting treatment options using approved therapy (7, 8).
Candida auris has now been identified in 28 U.S. states and will likely continue to spread within those states and to new areas over time (Centers for Disease Control and Prevention [CDC]; https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html). Health care transmission of C. auris tends to disproportionately impact high-acuity long-term-care facilities and individuals with chronic illness, history of other resistant pathogens, and invasive medical devices, including mechanical ventilation, tracheostomies, feeding tubes, and urinary catheters (9). C. auris is associated with high mortality, but because infected or colonized individuals often have poor health at baseline, the attributable mortality of C. auris is not clear (6). Colonized or infected individuals shed C. auris into their immediate environment, where it can persist for long periods of time on health care surfaces, including shared medical equipment, potentially spreading to others and causing health care outbreaks (5). Prompt identification of individuals infected or colonized with C. auris is critical for guiding infection control measures, including isolation precautions and thorough cleaning and disinfection of their surroundings to prevent spread. Infection control guidance can be found on the CDC website (https://www.cdc.gov/fungal/candida-auris/c-auris-infection-control.html), and similar recommendations have been published by the Australasian Society for Infectious Diseases and the International Society for Antimicrobial Chemotherapy (10, 11). The CDC has also issued additional guidance for working with C. auris in a clinical microbiology laboratory. The guidance can be found at https://www.cdc.gov/fungal/candida-auris/c-auris-lab-safety.html.
Because C. auris was a newly identified species when it began emerging in multiple countries across the world, it did not appear in any identification platform databases (12). To make matters worse, its biochemical assimilation profile was very similar to that of two closely related species, Candida haemulonii and Candida duobushaemulonii, causing a great deal of misidentification (13). At least one laboratory system identified C. auris isolates as Saccharomyces cerevisiae, and those laboratories with early versions of MALDI-TOF MS either identified it as C. haemulonii or did not get an identification at all (12, 13). Complicating the identification of C. auris is the fact that there are five different clades (including a few cases from clade V so far identified only in Iran) and the clades have both different assimilation profiles and slight differences in their rDNA sequences (2, 14).
BIOCHEMISTRY-BASED IDENTIFICATION OF CANDIDA AURIS
Many commercial identification platforms use biochemical assimilation and fermentation patterns to identify bacteria and fungi. This poses a problem for the identification of C. auris, as the assimilation and fermentation patterns are similar to those of other closely related species of yeast. Complicating the problem, some identification systems make an identification to species based on the closest match to assimilation and fermentation pattern by percent match, rather than based on a perfect match. While this may work for the 90 to 95% of Candida isolates that comprise the six most frequently identified species (C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, and C. lusitaniae), it can cause inaccurate identification of some of the other 5 to 10% of Candida species. Rather than giving “no identification,” they will make an erroneous identification. This limitation caused many of the first isolates of C. auris to be incorrectly identified as Rhodotorula glutinis, Saccharomyces cerevisiae, or Candida sake when the API 20C AUX or API ID 32C system was used or as Candida haemulonii when the Vitek 2 system was used (12, 15, 16). Efforts to incorporate C. auris into these databases have led to some incremental improvements. For example, although an update to the Vitek 2 database (version 8.01) enabled accurate identification of C. auris clade IV isolates, this update was not sufficient to consistently identify isolates from clade I or III (17). Vitek 2 has since released version 9.01, but an evaluation for improved inclusivity has not yet been published. Other identification platforms, such as the MicroScan Walkaway and the BD Phoenix, have not yet added C. auris to their databases and continue to provide erroneous identification (Table 1) (12, 18). For these reasons, the CDC has created algorithms to indicate when additional follow-up may be needed for a variety of commonly used biochemistry-based identification systems (https://www.cdc.gov/fungal/candida-auris/pdf/Testing-algorithm_by-Method_508.pdf).
TABLE 1.
Methods for identification or isolation of Candida auris
| Test type and details | Notesa | Reference(s) |
|---|---|---|
| Culture | ||
| Original enrichment broth | Valuable reference method for diagnostic development | 30 |
| Chromogenic medium | Aids visual identification to the species level of the common Candida spp. | 24, 26, 27 |
| Other differential media | Use of Pal’s medium, ferrous sulfate, and crystal violet | 25, 28, 29 |
| Biochemical tests | ||
| API 20C AUX | Cannot currently identify C. auris; see CDC follow-up algorithm | 12, 15, 16 |
| API ID 32C | Cannot currently identify C. auris; see CDC follow-up algorithm | 12 |
| BD Phoenix | Cannot currently identify C. auris; see CDC follow-up algorithm | 12 |
| MicroScan | Cannot currently identify C. auris; see CDC follow-up algorithm | 12 |
| RapID yeast plus | Cannot currently identify C. auris; see CDC follow-up algorithm | |
| Vitek 2 YST | Can ID some but not all C. auris; see CDC follow-up algorithm | 17 |
| MALDI-TOF MS | ||
| Bruker Biotyper 2.0 Microflex LT | FDA approved for isolate ID with CA System library (v4) | 20 |
| bioMérieux Vitek MS | FDA approved for isolate ID with IVD library v3.2 | 19 |
| Blood culture, molecular | ||
| BioFire BCID2 | FDA approved for positive blood culture | |
| GenMark Dx ePlex BCID-FP panel | FDA approved for positive blood culture | 58 |
| RT-PCR | ||
| TaqMan chemistry | Most common LDT for colonization screening in U.S. PHL | 41, 52 |
| SYBR green chemistry | Evaluated for skin and anterior nares | 39, 42 |
| Commercial RT-PCR kits | ||
| AurisID, OLM Diagnostics | CE-IVD reagents for C. auris RT-PCR | 47 |
| BioGX Candida auris | RUO reagents supporting RT-PCR and extraction on BD Max platform | |
| Fungiplex Candida auris | RUO reagents for C. auris RT-PCR | 47 |
| Other | ||
| LAMP | Unique molecular method for C. auris detection | 40 |
| T2MR C. auris | RUO test for C. auris using T2 magnetic resonance technology | 50 |
| Conventional PCR with GPI target | C. auris specific and multiplex tests feasible in low-resource settings | 36 – 38 |
ID, identification; LDT, laboratory-developed test; RUO, research use only; PHL, public health laboratories; CE-IVD, in vitro diagnostic approved for sale in the European Union; RT-PCR, real-time PCR.
MASS SPECTROMETRY-BASED IDENTIFICATION
As it was a new species and was not represented in the databases, initial attempts to identify C. auris based on matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) were unsuccessful. Following the continual isolation and spread of C. auris across many countries, the commercial manufacturers of MALDI-TOF MS added it first to their research use only (RUO) databases and then subsequently to their FDA-cleared databases (19, 20). Isolates from all four of the major C. auris clades can now be correctly identified with the Vitek MS system (bioMérieux, Durham, NC) by using its FDA-cleared IVD v3.2 or RUO Saramis v4.14 or newer databases and the Biotyper 2.0 Microflex LT spectrometer (Bruker Daltonics, Inc., Billerica, MA) by using the CA system library (version claim 4) or RUO version 2014 (5627) or newer databases.
IDENTIFICATION OF C. AURIS USING AGAR
Chromogenic medium has been a staple diagnostic tool for Candida species identification for a few decades (21). Candida species are identified based on colony color and, in the case of C. krusei, colony texture. There are many different commercially available formulations, but the majority claim identification for only C. albicans, C. tropicalis, C. krusei, and, depending on the formulation, C. glabrata (21–23). Most chromogenic medium formulations are not capable of reliably distinguishing C. auris, as colonies can appear cream, pink, red, or purple and resemble the majority of other species besides the four mentioned above (https://www.cdc.gov/fungal/candida-auris/identification.html) (24, 25). Recently, two new formulations of chromogenic media have been developed specifically for the additional identification of C. auris, CHROMagar Candida plus (CHROMagar, France) (Fig. 1) and HiCrome C. auris MDR selective agar (HiMedia, Mumbai, India) (24, 26, 27). When tested side by side against 49 Candida isolates including representatives from all four major C. auris clades, only CHROMagar Candida plus correctly distinguished all the C. auris isolates (24). However, with CHROMagar Candida plus, there were false-positive identifications with the closely related species Candida vulturna and Candida pseudohaemulonii. For this reason, colonies suspected of being C. auris by using colony color on chromogenic media should be further confirmed by sequencing or MALDI-TOF MS (24).
FIG 1.
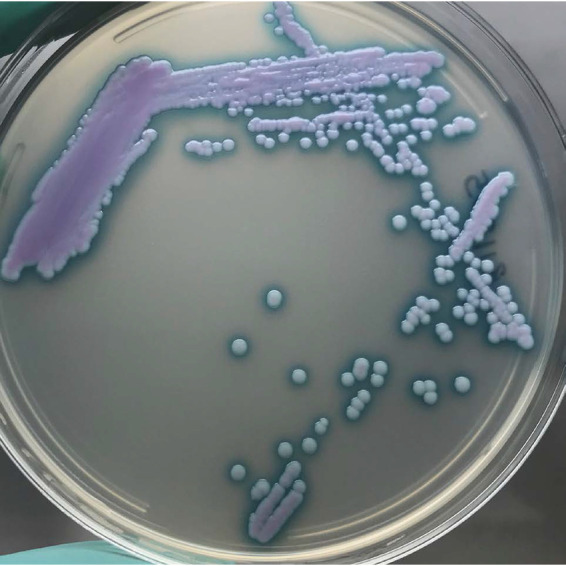
Candida auris after 48 h of growth on CHROMagar Candida plus showing light blue colonies with a blue halo around the colonies. The combination of the color and the halo are distinct for C. auris (also see reference 22).
The use of Pal’s medium in combination with CHROMagar has been suggested as a way to distinguish between the closely related species C. auris and species in the C. haemulonii species complex, but this option is useful only when an identification has been narrowed down to these closely related species (28).
Two other noncommercial selective agars for C. auris have been recently developed (25, 29). The first medium uses high NaCl (12.5%) and ferrous sulfate in combination with elevated temperatures to allow only the differential growth of C. auris. This medium can be used to isolate C. auris when directly plated from blood culture as well (25). The second medium, developed by Ibrahim et al., is an alternative enrichment broth that uses 10% NaCl, mannitol, and crystal violet to differentially select for C. auris growth (29, 30). Both media will need to be validated against more Candida species, including closely related species in the C. haemulonii complex.
MOLECULAR IDENTIFICATION
DNA sequencing played a major role first in the initial discovery that C. auris was a new species and then in the correct identification of C. auris and its delineation from the closely related species in the C. haemulonii species complex (15, 31, 32). Identification can be accomplished by PCR amplification of the D1/D2 region of the 28S sequence or the full internal transcribed spacer of the ribosomal cistron (1). Unfortunately, DNA sequencing is not available in most clinical microbiology laboratories, and the majority of sequencing is performed in reference laboratories.
Conventional PCR, using species-specific primers and/or restriction length polymorphism analysis, for the identification of Candida species has been in use for decades (33–35). Due to the availability of both cheaper and easier to use commercial methods, this methodology is not widely used outside of research or reference laboratories and has not been commercialized. However, in resource-limited settings, conventional PCR can be used to identify specific emerging species such as C. auris (36–38). The methodology can be incorporated into an algorithm in which chromogenic medium is used to screen isolates for possible C. auris and traditional PCR is used to confirm the identity.
A number of real-time PCR assays and a loop-mediated isothermal amplification (LAMP) assay for C. auris have now been developed using many different DNA extraction methods, enzyme chemistries, primer targets, and thermocycler platforms (36, 39–46). Each of these methods has been optimized for use in the laboratory in which they were developed, but they provide a variety of options for clinical laboratories that have the ability to incorporate laboratory-developed tests. Commercially available kits that include premixed primers and enzymes are also now available, such as the AurisID (OLM Diagnostics, Newcastle Upon Tyne, United Kingdom) and the Fungiplex (Bruker, Bremen, Germany), both of which can be used for detection from surveillance specimens or directly from blood, and the BioGX Candida auris kit (BioGX, Birmingham, AL) for the BD Max (Becton, Dickinson and Company, Franklin Lakes, NJ) (47–49). Other unique molecular tests include the T2 magnetic resonance (MR) assay, which was originally developed for blood specimens but has also been adapted for colonization screening (50). The T2Cauris panel is currently being marketed for research use only and has not been cleared for diagnostic use. In the United States, the FDA has approved both the GenMark Dx ePlex BCID-FP panel and BioFire’s BCID 2 panel for the detection of C. auris in positive blood cultures.
DETECTION OF COLONIZATION
Another aspect of laboratory testing for C. auris is the use of screening to determine whether patients may be colonized with C. auris. As stated above, C. auris primarily colonizes the skin, is shed into the health care environment, where it contaminates surfaces including shared medical equipment, and is easily transferred between patients. For this reason, appropriate infection control precautions should be used for patients colonized or infected with C. auris to prevent further spread. Screening using skin swabs is useful for identifying asymptomatically colonized individuals and is often used for patients at high risk for acquiring C. auris, including health care contacts of known cases, patients with domestic or international health care in an area with C. auris, or patients with current or previous stays at long-term-care facilities. Some health care facilities will screen individuals upon admission or conduct point prevalence surveys in which all patients in the facility or a specific unit are screened. There are several laboratory-developed methods for screening patients for C. auris colonization, including both a conventional broth enrichment method and a number of real-time PCR methods. Rapid and easy methods for colonization screening are important, because results are used to guide infection control measures, e.g., whether a patient requires isolation precautions.
Broth culture was the first method developed for screening for C. auris colonization (30). Sample collection involves bilateral swabbing of the axilla and groin of patients to be screened, followed by enrichment for C. auris in Sabouraud broth containing 10% salt and dulcitol (30). A brief protocol for this methodology has been published (51), but the CDC has made a standard operating procedure for the method from start to finish available online (https://www.cdc.gov/fungal/lab-professionals/pdf/c.-auris-colonization-screening-508.pdf). Additional guidance for swabbing patients can be found at https://www.cdc.gov/fungal/candida-auris/c-auris-patient-swab.html.
At the time of this writing, there was no FDA-approved test specific for C. auris colonization swab specimen screening, an existing gap in diagnostics. Because the public health response is oriented around colonization screening for early detection and containment, colonization screening demands the highest testing volumes by far and capacity is often limited. In the United States, most public health laboratories capable of C. auris colonization screening have validated a version of a TaqMan real-time PCR (RT-PCR) assay first developed by the Wadsworth Center in New York, run on either the ABI 7500 thermocycler or the BD Max platform (41, 52).
Current public health surveillance testing in the United States is managed through the Antimicrobial Resistance Laboratory Network (AR Lab Network) (https://www.cdc.gov/drugresistance/laboratories.html) and coordinated through state and jurisdictional public health laboratories. There are some clinical and private laboratories that perform targeted colonization screening of newly admitted patients, but wide-scale surveillance testing and large point prevalence surveys are largely performed through the AR Lab Network. Candida auris is a reportable disease in the United States, and positive cases should be reported through state and jurisdictional public health laboratories but can also be coordinated through notification at candidaauris@cdc.gov.
SUSCEPTIBILITY TESTING
One of the more worrisome attributes of C. auris is the percentage of isolates that are antimicrobial resistant (53). There are no breakpoints for any antimicrobials against C. auris. While MIC values can be generated using commercially available susceptibility testing platforms, there are no interpretive criteria. As there are no clinical trials of currently available antimicrobials against C. auris, there are not likely to be established breakpoints in the near future either. Many laboratories refer to the tentative breakpoints that were established by the CDC based on MIC distributions, animal models of infection, and molecular identification of resistance mechanisms (https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html) (2, 54). While these tentative breakpoints may assist clinicians with MIC interpretation, they are not endorsed by the Clinical and Laboratory Standards Institute (CLSI), and these interpretations should not be included in patient reports.
There are additional concerns with susceptibility and C. auris. A high number of isolates exhibit the Eagle effect when testing using broth microdilution against caspofungin (13, 55, 56). If other echinocandins are not tested simultaneously with caspofungin, this could lead to the major error of predicting echinocandin resistance in a susceptible isolate. In addition, amphotericin B testing can be problematic. As the CLSI has not established amphotericin B breakpoints for any fungal species, most laboratories use a value of ≥2 μg/mL as the breakpoint for resistance to amphotericin B for all Candida species. Using broth microdilution, the majority of Candida isolates have MIC values of 0.25 to 1 μg/mL. If an isolate is read just 1 or 2 dilutions higher at 2 μg/mL, the two readings are in essential agreement, within the ±2 dilution standard deviation, and still in categorical disagreement. Since reading is subjective and MIC values can fluctuate within the standard deviation, there are challenges with performing amphotericin B susceptibility testing. The use of gradient diffusion increases the range of obtainable MIC values, but since it is also a subjectively read test, isolates hovering between resistant and susceptible are difficult to interpret.
TESTING STRATEGIES
In a resource reference laboratory, MALDI-TOF MS or DNA sequencing should be used first for the identification of isolates that are suspected to be Candida auris. If neither of these options are available, the Vitek 2 system is the next most accurate way to identify isolates of C. auris, but the accuracy is not as high as for MALDI-TOF MS or sequencing, and some clades may be identified as members of the C. haemulonii species complex. Those isolates may have to be characterized further. When none of these options are available, CHROMagar Candida plus can be used to identify isolates by color. The specificity of this agar is not 100%, so for isolates that have a preliminary identification of C. auris, a species-specific PCR or RT-PCR assay should be used to confirm the identity. If none of these options are available, the isolate should be forwarded to a reference laboratory for identification.
ADDITIONAL TOOLS
Isolates of C. auris from all five clades for validation or study have been made available for free from the CDC AR Isolate Bank. The AR Isolate Bank and all available isolates can be found at https://www.cdc.gov/drugresistance/resistance-bank/index.html.
Whole-genome sequencing (WGS) is quickly becoming a useful tool for outbreak investigation and surveillance. While WGS is more difficult with a eukaryote like C. auris, there are already several labs that have incorporated it into their surveillance. To that end, the CDC has made a benchmark data set of 23 C. auris genomes available, and the directions for finding them and using them can be found in a report by Welsh et al. (57). In addition, there are publicly available data analysis tools (https://github.com/CDCgov/mycosnp), and the CDC has established a C. auris umbrella project on the NCBI website (NCBI accession no. PRJNA642852).
SUMMARY
As more laboratories have focused on the identification of C. auris, the number of available tools for detection and identification has increased. However, not all laboratories can implement a laboratory-developed test, and many do not have access to MALDI-TOF MS or a Vitek 2 system. The number of alternative commercially available tests is still quite limited, which leaves definitive identification of C. auris as a send-out test in many facilities. An especially glaring deficiency is the lack of a point-of-care test for detection of C. auris colonization. This would be an important tool for the identification of colonized patients, which plays an important role in the implementation of infection control practices, especially in cities where C. auris has become endemic. Candida auris continues to spread across the United States and across the world, and it has become a notifiable disease in many U.S. states and municipalities. While we may only be able to slow the spread, more tools for its detection and identification will allow us to combat it at the point of individual patient care.
ACKNOWLEDGMENT
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Contributor Information
Shawn R. Lockhart, Email: gyi2@cdc.gov.
Romney M. Humphries, Vanderbilt University Medical Center
REFERENCES
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lockhart SR, Berkow EL, Chow N, Welsh RM. 2017. Candida auris for the clinical microbiology laboratory: not your grandfather's Candida species. Clin Microbiol Newsl 39:99–103. 10.1016/j.clinmicnews.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proctor DM, Dangana T, Sexton DJ, Fukuda C, Yelin RD, Stanley M, Bell PB, Baskaran S, Deming C, Chen Q, Conlan S, Park M, Welsh RM, Vallabhaneni S, Chiller T, Forsberg K, Black SR, Pacilli M, Kong HH, Lin MY, Schoeny ME, Litvintseva AP, Segre JA, Hayden MK, NISC Comparative Sequencing Program. 2021. Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat Med 27:1401–1409. 10.1038/s41591-021-01383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sexton DJ, Bentz ML, Welsh RM, Derado G, Furin W, Rose LJ, Noble-Wang J, Pacilli M, McPherson TD, Black S, Kemble SK, Herzegh O, Ahmad A, Forsberg K, Jackson B, Litvintseva AP. 2021. Positive correlation between Candida auris skin-colonization burden and environmental contamination at a ventilator-capable skilled nursing facility in Chicago. Clin Infect Dis 73:1142–1148. 10.1093/cid/ciab327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams E, Quinn M, Tsay S, Poirot E, Chaturvedi S, Southwick K, Greenko J, Fernandez R, Kallen A, Vallabhaneni S, Haley V, Hutton B, Blog D, Lutterloh E, Zucker H, Candida auris Investigation Workgroup. 2018. Candida auris in healthcare facilities, New York, USA, 2013–2017. Emerg Infect Dis 24:1816–1824. 10.3201/eid2410.180649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrowsky B, Greenko J, Adams E, Quinn M, O'Brien B, Chaturvedi V, Berkow E, Vallabhaneni S, Forsberg K, Chaturvedi S, Lutterloh E, Blog D, C. auris Investigation Work Group. 2020. Candida auris isolates resistant to three classes of antifungal medications—New York, 2019. MMWR Morb Mortal Wkly Rep 69:6–9. 10.15585/mmwr.mm6901a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyman M, Forsberg K, Reuben J, Dang T, Free R, Seagle EE, Sexton DJ, Soda E, Jones H, Hawkins D, Anderson A, Bassett J, Lockhart SR, Merengwa E, Iyengar P, Jackson BR, Chiller T. 2021. Notes from the field: transmission of pan-resistant and echinocandin-resistant Candida auris in health care facilities—Texas and the District of Columbia, January-April 2021. MMWR Morb Mortal Wkly Rep 70:1022–1023. 10.15585/mmwr.mm7029a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossow J, Ostrowsky B, Adams E, Greenko J, McDonald R, Vallabhaneni S, Forsberg K, Perez S, Lucas T, Alroy KA, Jacobs Slifka K, Walters M, Jackson BR, Quinn M, Chaturvedi S, Blog D, New York Candida auris Investigation Workgroup. 2021. Factors associated with Candida auris colonization and transmission in skilled nursing facilities with ventilator units, New York, 2016–2018. Clin Infect Dis 72:e753–e760. 10.1093/cid/ciaa1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong CW, Chen SC, Clark JE, Halliday CL, Kidd SE, Marriott DJ, Marshall CL, Morris AJ, Morrissey CO, Roy R, Slavin MA, Stewardson AJ, Worth LJ, Heath CH, Australian and New Zealand Mycoses Interest Group (ANZMIG), Healthcare Infection Control Special Interest Group (HICSIG); both of the Australasian Society for Infectious Diseases (ASID). 2019. Diagnosis, management and prevention of Candida auris in hospitals: position statement of the Australasian Society for Infectious Diseases. Intern Med J 49:1229–1243. 10.1111/imj.14612. [DOI] [PubMed] [Google Scholar]
- 11.Kenters N, Kiernan M, Chowdhary A, Denning DW, Pemán J, Saris K, Schelenz S, Tartari E, Widmer A, Meis JF, Voss A. 2019. Control of Candida auris in healthcare institutions: outcome of an International Society for Antimicrobial Chemotherapy expert meeting. Int J Antimicrob Agents 54:400–406. 10.1016/j.ijantimicag.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, Hewitt C, Simner PJ, Carroll KC, Hayden RT, Zhang SX. 2017. Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 55:638–640. 10.1128/JCM.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prakash A, Sharma C, Singh A, Kumar Singh P, Kumar A, Hagen F, Govender NP, Colombo AL, Meis JF, Chowdhary A. 2016. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect 22:277.e1-9. 10.1016/j.cmi.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 15.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49:3139–3142. 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowdhary A, Anil Kumar V, Sharma C, Prakash A, Agarwal K, Babu R, Dinesh KR, Karim S, Singh SK, Hagen F, Meis JF. 2014. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33:919–926. 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 17.Ambaraghassi G, Dufresne PJ, Dufresne SF, Vallières É, Muñoz JF, Cuomo CA, Berkow EL, Lockhart SR, Luong ML. 2019. Identification of Candida auris by use of the updated Vitek 2 yeast identification system, version 8.01: a multilaboratory evaluation study. J Clin Microbiol 57:e00884-19. 10.1128/JCM.00884-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snayd M, Dias F, Ryan RW, Clout D, Banach DB. 2018. Misidentification of Candida auris by RapID Yeast Plus, a commercial, biochemical enzyme-based manual rapid identification system. J Clin Microbiol 56:e00080-18. 10.1128/JCM.00080-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girard V, Mailler S, Chetry M, Vidal C, Durand G, van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. 2016. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses 59:535–538. 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 20.Voelker R. 2018. New test identifies Candida auris. JAMA 319:2164. 10.1001/jama.2018.6922. [DOI] [PubMed] [Google Scholar]
- 21.Odds FC, Bernaerts R. 1994. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol 32:1923–1929. 10.1128/jcm.32.8.1923-1929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaller MA, Houston A, Coffmann S. 1996. Application of CHROMagar Candida for rapid screening of clinical specimens for Candida albicans, Candida tropicalis, Candida krusei, and Candida (Torulopsis) glabrata. J Clin Microbiol 34:58–61. 10.1128/jcm.34.1.58-61.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernal S, Martín Mazuelos E, García M, Aller AI, Martínez MA, Gutiérrez MJ. 1996. Evaluation of CHROMagar Candida medium for the isolation and presumptive identification of species of Candida of clinical importance. Diagn Microbiol Infect Dis 24:201–204. 10.1016/0732-8893(96)00063-6. [DOI] [PubMed] [Google Scholar]
- 24.de Jong AW, Dieleman C, Carbia M, Mohd Tap R, Hagen F. 2021. Performance of two novel chromogenic media for the identification of multidrug-resistant Candida auris compared with other commercially available formulations. J Clin Microbiol 59:e03220-20. 10.1128/JCM.03220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das S, Singh S, Tawde Y, Chakrabarti A, Rudramurthy SM, Kaur H, Shankarnarayan SA, Ghosh A. 2021. A selective medium for isolation and detection of Candida auris, an emerging pathogen. J Clin Microbiol 59:e00326-20. 10.1128/JCM.00326-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borman AM, Fraser M, Johnson EM. 2021. CHROMagarTM Candida Plus: a novel chromogenic agar that permits the rapid identification of Candida auris. Med Mycol 59:253–258. 10.1093/mmy/myaa049. [DOI] [PubMed] [Google Scholar]
- 27.Mulet Bayona JV, Salvador García C, Tormo Palop N, Gimeno Cardona C. 2020. Evaluation of a novel chromogenic medium for Candida spp. identification and comparison with CHROMagar™ Candida for the detection of Candida auris in surveillance samples. Diagn Microbiol Infect Dis 98:115168. 10.1016/j.diagmicrobio.2020.115168. [DOI] [PubMed] [Google Scholar]
- 28.Kumar A, Sachu A, Mohan K, Vinod V, Dinesh K, Karim S. 2017. Simple low cost differentiation of Candida auris from Candida haemulonii complex using CHROMagar Candida medium supplemented with Pal's medium. Rev Iberoam Micol 34:109–111. 10.1016/j.riam.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Ibrahim A, Peyclit L, Abdallah R, Khelaifia S, Chamieh A, Rolain JM, Bittar F. 2021. SCA medium: a new culture medium for the isolation of all Candida auris clades. J Fungi (Basel) 7:433. 10.3390/jof7060433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, Litvintseva AP. 2017. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol 55:2996–3005. 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19:1670–1673. 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magobo RE, Corcoran C, Seetharam S, Govender NP. 2014. Candida auris-associated candidemia, South Africa. Emerg Infect Dis 20:1250–1251. 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niesters HG, Goessens WH, Meis JF, Quint WG. 1993. Rapid, polymerase chain reaction-based identification assays for Candida species. J Clin Microbiol 31:904–910. 10.1128/jcm.31.4.904-910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jordan JA. 1994. PCR identification of four medically important Candida species by using a single primer pair. J Clin Microbiol 32:2962–2967. 10.1128/jcm.32.12.2962-2967.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schönian G, Meusel O, Tietz HJ, Meyer W, Gräser Y, Tausch I, Presber W, Mitchell TG. 1993. Identification of clinical strains of Candida albicans by DNA fingerprinting with the polymerase chain reaction. Mycoses 36:171–179. 10.1111/j.1439-0507.1993.tb00746.x. [DOI] [PubMed] [Google Scholar]
- 36.Alvarado M, Bartolomé Álvarez J, Lockhart SR, Valentín E, Ruiz-Gaitán AC, Eraso E, de Groot PWJ. 2021. Identification of Candida auris and related species by multiplex PCR based on unique GPI protein-encoding genes. Mycoses 64:194–202. 10.1111/myc.13204. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz-Gaitán AC, Fernández-Pereira J, Valentin E, Tormo-Mas MA, Eraso E, Pemán J, de Groot PWJ. 2018. Molecular identification of Candida auris by PCR amplification of species-specific GPI protein-encoding genes. Int J Med Microbiol 308:812–818. 10.1016/j.ijmm.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 38.Theill L, Dudiuk C, Morales-Lopez S, Berrio I, Rodríguez JY, Marin A, Gamarra S, Garcia-Effron G. 2018. Single-tube classical PCR for Candida auris and Candida haemulonii identification. Rev Iberoam Micol 35:110–112. 10.1016/j.riam.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Georgacopoulos O, Nunnally NS, Le N, Lysen C, Welsh RM, Kordalewska M, Perlin DS, Berkow EL, Sexton DJ. 2020. Performance evaluation of culture-independent SYBR green Candida auris quantitative PCR diagnostics on anterior nares surveillance swabs. J Clin Microbiol 58:e00690-20. 10.1128/JCM.00690-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto M, Alshahni MM, Tamura T, Satoh K, Iguchi S, Kikuchi K, Mimaki M, Makimura K. 2018. Rapid detection of Candida auris based on loop-mediated isothermal amplification (LAMP). J Clin Microbiol 56:e00591-18. 10.1128/JCM.00591-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leach L, Zhu Y, Chaturvedi S. 2018. Development and validation of a real-time PCR assay for rapid detection of Candida auris from surveillance samples. J Clin Microbiol 56:e01223-17. 10.1128/JCM.01223-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, Perlin DS. 2017. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol 55:2445–2452. 10.1128/JCM.00630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walchak RC, Buckwalter SP, Zinsmaster NM, Henn KM, Johnson KM, Koelsch JM, Herring SA, Steinmetz LK, Reed KA, Barth JE, Rasmusson JM, Fischer JL, Snippes VP, Sampathkumar P, Wengenack NL. 2020. Candida auris direct detection from surveillance swabs, blood, and urine using a laboratory-developed PCR method. J Fungi (Basel) 6:224. 10.3390/jof6040224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malczynski M, Dowllow N, Rezaeian S, Rios J, Dirnberger L, Zembower JA, Zhu A, Qi C. 2020. Optimizing a real-time PCR assay for rapid detection of Candida auris in nasal and axillary/groin samples. J Med Microbiol 69:824–829. 10.1099/jmm.0.001207. [DOI] [PubMed] [Google Scholar]
- 45.Arastehfar A, Fang W, Daneshnia F, Al-Hatmi AM, Liao W, Pan W, Khan Z, Ahmad S, Rosam K, Lackner M, Lass-Flörl C, Hagen F, Boekhout T. 2019. Novel multiplex real-time quantitative PCR detecting system approach for direct detection of Candida auris and its relatives in spiked serum samples. Future Microbiol 14:33–45. 10.2217/fmb-2018-0227. [DOI] [PubMed] [Google Scholar]
- 46.Ibrahim A, Baron SA, Yousfi H, Hadjadj L, Lalaoui R, Morand S, Rolain JM, Bittar F. 2021. Development and standardization of a specific real-time PCR assay for the rapid detection of Candida auris. Eur J Clin Microbiol Infect Dis 40:1547–1551. 10.1007/s10096-021-04176-8. [DOI] [PubMed] [Google Scholar]
- 47.Sattler J, Noster J, Brunke A, Plum G, Wiegel P, Kurzai O, Meis JF, Hamprecht A. 2021. Comparison of two commercially available qPCR kits for the detection of Candida auris. J Fungi (Basel) 7:154. 10.3390/jof7020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lima A, Widen R, Vestal G, Uy D, Silbert S. 2019. A TaqMan probe-based real-time PCR assay for the rapid identification of the emerging multidrug-resistant pathogen Candida auris on the BD Max system. J Clin Microbiol 57:e01604-18. 10.1128/JCM.01604-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulet Bayona JV, Salvador García C, Tormo Palop N, Gimeno Cardona C. 2021. Validation and implementation of a commercial real-time PCR assay for direct detection of Candida auris from surveillance samples. Mycoses 64:612–615. 10.1111/myc.13250. [DOI] [PubMed] [Google Scholar]
- 50.Sexton DJ, Bentz ML, Welsh RM, Litvintseva AP. 2018. Evaluation of a new T2 magnetic resonance assay for rapid detection of emergent fungal pathogen Candida auris on clinical skin swab samples. Mycoses 61:786–790. 10.1111/myc.12817. [DOI] [PubMed] [Google Scholar]
- 51.Bentz ML, Sexton DJ, Welsh RM, Litvintseva AP. 2018. Phenotypic switching in newly emerged multidrug-resistant pathogen Candida auris. Med Mycol 10.1093/mmy/myy100. [DOI] [PubMed] [Google Scholar]
- 52.Leach L, Russell A, Zhu Y, Chaturvedi S, Chaturvedi V. 2019. A rapid and automated sample-to-result Candida auris real-time PCR assay for high-throughput testing of surveillance samples with the BD Max open system. J Clin Microbiol 57:e00630-19. 10.1128/JCM.00630-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lockhart SR. 2019. Candida auris and multidrug resistance: defining the new normal. Fungal Genet Biol 131:103243. 10.1016/j.fgb.2019.103243. [DOI] [PubMed] [Google Scholar]
- 54.Lepak AJ, Zhao M, Berkow EL, Lockhart SR, Andes DR. 2017. Pharmacodynamic optimization for treatment of invasive Candida auris infection. Antimicrob Agents Chemother 61:e00791-17. 10.1128/AAC.00791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kordalewska M, Lee A, Park S, Berrio I, Chowdhary A, Zhao Y, Perlin DS. 2018. Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob Agents Chemother 62:e00238-18. 10.1128/AAC.00238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudramurthy SM, Chakrabarti A, Paul RA, Sood P, Kaur H, Capoor MR, Kindo AJ, Marak RSK, Arora A, Sardana R, Das S, Chhina D, Patel A, Xess I, Tarai B, Singh P, Ghosh A. 2017. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J Antimicrob Chemother 72:1794–1801. 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 57.Welsh RM, Misas E, Forsberg K, Lyman M, Chow NA. 2021. Candida auris whole-genome sequence benchmark dataset for phylogenomic pipelines. J Fungi (Basel) 7:214. 10.3390/jof7030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang SX, Carroll KC, Lewis S, Totten M, Mead P, Samuel L, Steed LL, Nolte FS, Thornberg A, Reid JL, Whitfield NN, Babady NE. 2020. Multicenter evaluation of a PCR-based digital microfluidics and electrochemical detection system for the rapid identification of 15 fungal pathogens directly from positive blood cultures. J Clin Microbiol 58:e02096-19. 10.1128/JCM.02096-19. [DOI] [PMC free article] [PubMed] [Google Scholar]


