Figure 4.
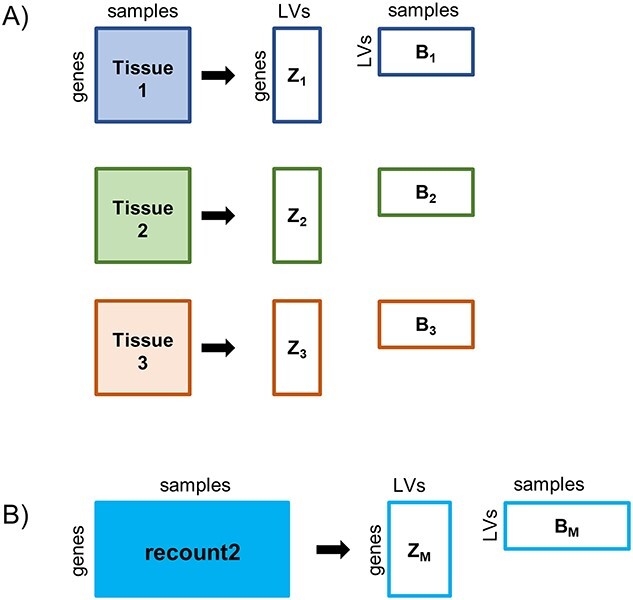
Overview of MultiPLIER framework. (A) PLIER [38], on which MultiPLIER is based, can analyze tissue-specific expression data and extract LVs by matrix factorization, resulting in matrices B and Z. PLIER then aligns the LVs with curated pathway gene sets in a downstream analysis. (B) To analyze data irrespective of tissue, MultiPLIER trains on a large collection of uniformly processed data in the form of the recount2 compendium [39], which contains around 370 000 samples. The resulting LVs can then be used to interpret a new dataset, by projecting the new gene expression data onto the latent space, to identify pathway-annotated LVs also featured in that new dataset. Diagram is based on [15].
