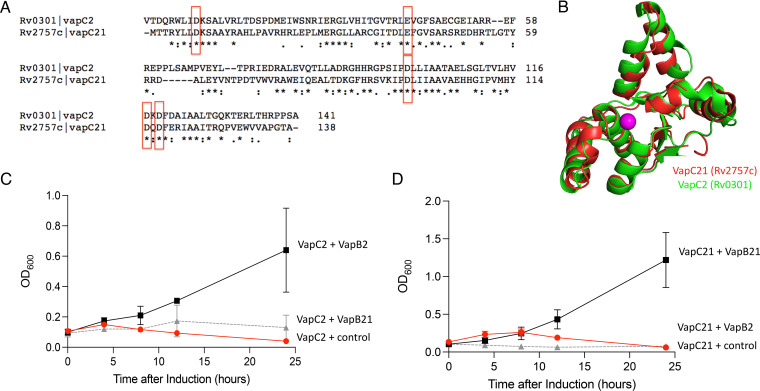
FIG 6.
VapC2 and VapC21 toxins are structurally similar, yet the VapB2 and VapB21 antitoxins are not interchangeable. (A) VapC2 and VapC21 sequence alignments were generated using Clustal Omega v 1.2.4 (45). The five conserved residues comprising the PIN domain are boxed in red. (B) Crystal structures of M. tuberculosis VapC2 (PDB 3H87) and VapC21 (PDB 5SV2) were superimposed using PyMOL. (C) Growth profiles of M. smegmatis carrying VapC2 toxin in pMC1s plasmid with either control pNIT vector (red circle), VapB2 antitoxin (black square), or VapB21 antitoxin (gray triangle). Growth curves performed in triplicate; standard deviation indicated. (D) Growth profiles of M. smegmatis carrying VapC21 toxin in pMC1s plasmid with either control pNIT vector (red circle), VapB21 antitoxin (black square), or VapB2 antitoxin (gray triangle).