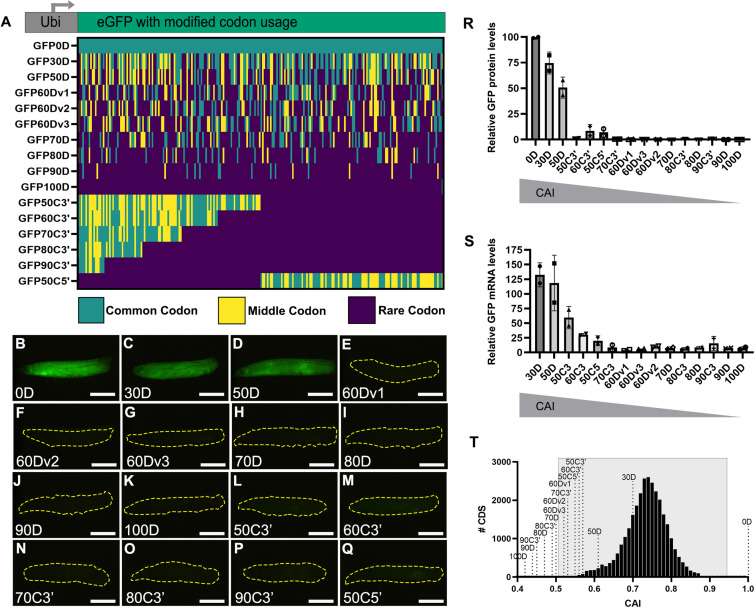
Figure 1. Strictly defined limits on rare codon usage regulate gene expression.
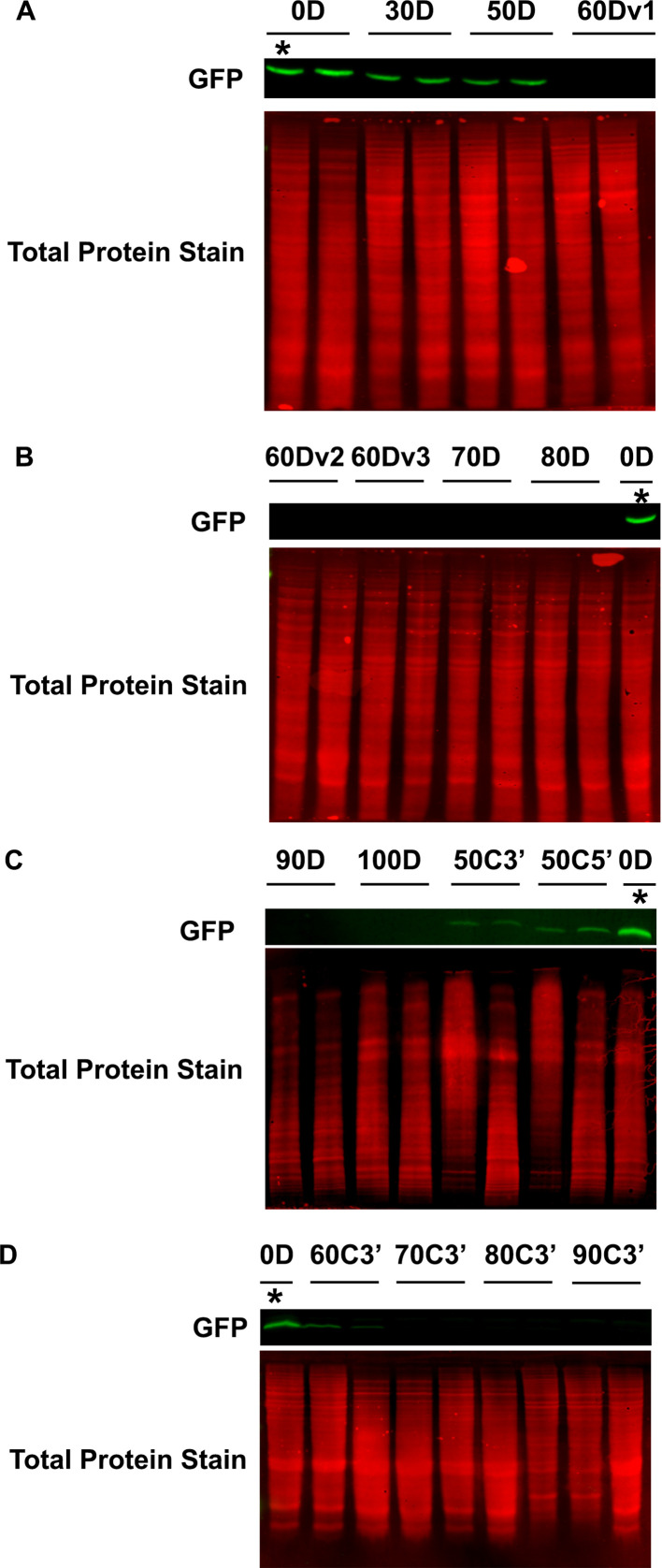
(A) Top- schematic of each indicated reporter. Bottom- heat map indicating codon usage across each reporter coding sequence. Color key indicates codon usage along the length of the coding sequence. (B–Q) Representative fluorescent images taken under identical camera exposure settings of live male wandering third instar larvae (WL3) containing stable genomic insertions of the indicated GFP reporter. Scalebars are 1 mm. Yellow dashed outlines highlight the larvae in images where no GFP signal was apparent. (R) Western blot protein quantifications of indicated reporters normalized to total protein stain then plotted as percentages relative to GFP0D for whole third instar larvae (two replicates, N=5 each, plotting individual data points and mean ± SEM). Reporters listed in descending order by Codon Adaptation Index (CAI). See Figure 1—figure supplement 1 for blot images. (S) mRNA abundances of the indicated reporters normalized as percentages relative to GFP0D for whole third instar larvae (two replicates, N=5 each, plotting individual data points and mean ± SEM). (T) Histogram of CAI values for each transcript in the Drosophila melanogaster genome. Reporter CAI values are indicated with dotted lines. Full range of endogenous genes highlighted in light gray box.