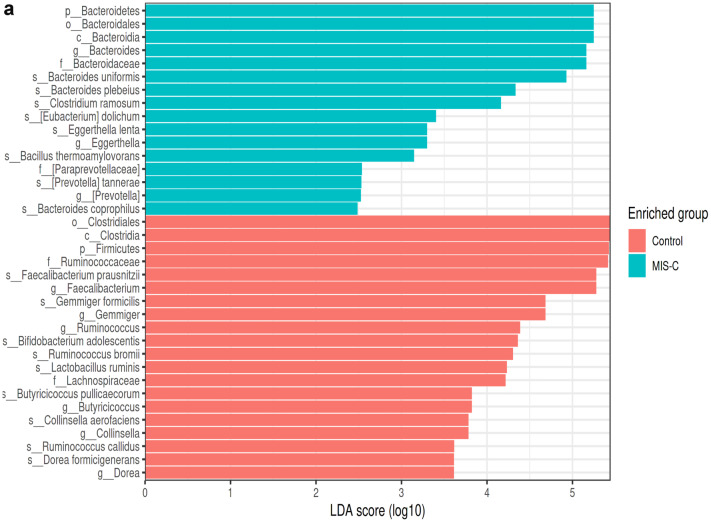
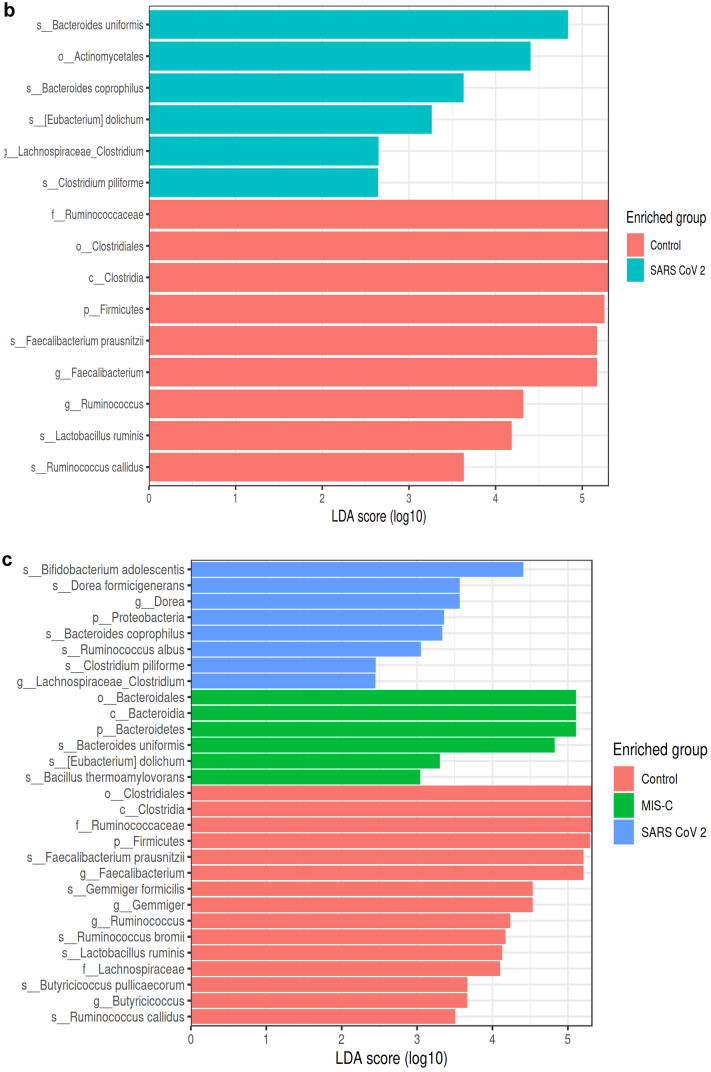
Fig. 4.
LEfSe analysis graphics showing bacterial taxa that were significantly different in abundance between the study groups. a–c LefSe analysis was performed to identify differentially abundant taxa for which the LDA scores are shown. Only species and functional modules with LDA effect size > 2 and FDR-corrected p value < 0.05 were plotted. Horizontal bars represent log.10 converted LDA score indicated by vertical dotted lines. Different colors represent different groups. p—phylum, c—class, o—order, f—family, g—genus, s—species. LEfSe analysis in stool samples for the MIS-C and control groups (control group (red), MIS-C (green)). a At the genus level, in the MIS-C group, Bacteroides (LDA score = 5.162, p = 0.018), Eggerthella (LDA score = 3.297, p = 0.017), and Prevotella (LDA score = 2.514, p = 0.038) were dominant, whereas Faecalibacterium (LDA score = 5.265, p = 0.0008), Gemmiger (LDA score = 4.666, p = 0.012), Ruminococcus (LDA score = 4.368, p = 0.000), Butyricicoccus (LDA score = 3.815, p = 0.002), Collinsella (LDA score = 3.752, p = 0.045), and Dorea (LDA score = 3.608, p = 0.045) were found to be dominant in the control group. At the species level, Bacteroides uniformis (LDA score = 4.926, p = 0.023), Bacteroides plebeius (LDA score = 4.332, p = 0.046), Clostridium ramosum (LDA score = 4.160, p = 0.046), Eubacterium dolichum (LDA score = 3.401, p = 0.023), Eggerthella lenta (LDA score = 3.296, p = 0.032), Bacillus thermoaamylovorans (LDA score = 3.146, p = 0.023), Prevotella tannerae (LDA score = 2.523, p = 0.038), and Bacteroides coprophilus (LDA score = 2.460, p = 0.041) are most abundant in the MIS-C group. In the control group, Faecalibacterium prausnitzii (LDA score = 5.265, p = 0.000), Gemmiger formicilis (LDA score = 4.666, p = 0.012), Bifidobacterium adolescentis (LDA score = 4.344, p = 0.008), Ruminococcus bromii (LDA score = 4.286, p = 0.000), Lactobacillus ruminis (LDA score = 4.231, p = 0.046), Butyricicoccus pullicaerocum (LDA score = 3.815, p = 0.002), Collinsella aerofaciens (LDA score = 3.752, p = 0.045), Ruminococcus callidus (LDA score = 3.609, p = 0.008), and Dorea formicigenerans (LDA score = 3.608, p = 0.000) were found to be dominant (a). LEfSe analysis in stool samples for the SARS-CoV-2 and control groups (control group (red), SARS-CoV-2 (blue)). b At the genus level, Lachnospiraceae_Clostridium (LDA score = 2.648, p = 0.039) was dominant in SARS-CoV-2 cases, while Faecalibacterium (LDA score = 5.168, p = 0.016) and Ruminococcus (LDA score = 4.316, p = 0.037) were dominant in the control group. At the species level, in the SARS-CoV-2 group, Bacteroides coprophilus (LDA score = 3.629, p = 0.002), Eubacterium dolichum (LDA score = 3.261, p = 0.012), Bacteroides uniformis (LDA score = 4.834, p = 0.011), and Clostridium piliforme (LDA score = 2.642, p = 0.039) were dominant, while Faecalibacterium prausnitzii (LDA score = 5.168, p = 0.016), Ruminococcus callidus (LDA score = 3.632, p = 0.003), and Lactobacillus ruminis (LDA score = 4.184, p = 0.013) were dominant in the control group (b). LEfSe analysis in stool samples for the MIS-C, SARS-CoV-2, and control groups (control group (red), MIS-C group (green), SARS-CoV-2 (blue)). c Bacteroides uniformis (LDA score = 4.820, p = 0.024), Bacillus thermoaamylovorans (LDA score = 3.040, p = 0.048), and Eubacterium dolichum (LDA score = 3.297, p = 0.002) were most abundant species in children with MIS-C at species level. Bacteroides coprophilus (LDA score = 3.331, p = 0.008), Bifidobacterium adolescentis (LDA score = 4.408, p = 0.016), Dorea formicigenerans (LDA score = 3.560, p = 0.000), Ruminococcus albus (LDA score = 3.049, p = 0.017), and Clostridium piliforme (LDA score = 2.450, p = 0.0130) were most abundant species in children with SARS-COV-2. In the control group, Faecalibacterium prausnitzii (LDA score = 5.201, p = 0.000), Gemmiger formicilis (LDA score = 4.524, p = 0.019), Ruminococcus bromii (LDA score = 4.169, p = 0.000), Lactobacillus ruminis (LDA score = 4.231, p = 0.046), Butyricicoccus pullicaerocum (LDA score = 3.660, p = 0.002), and Ruminococcus callidus (LDA score = 3.499, p = 0.000) were found to be dominant (c)