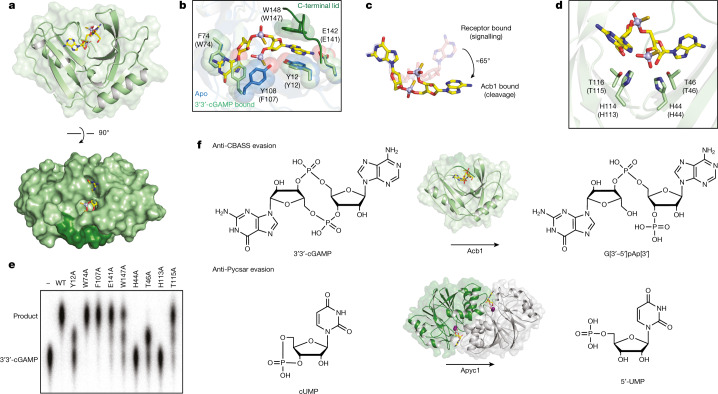
Fig. 3. Structural basis of Acb1 3′3′-cGAMP degradation.
a, Overview of Acb1 from Erwinia phage FBB1 in complex with a hydrolysis-resistant phosphorothioate analogue of 3′3′-cGAMP. In the surface representation, C-terminal lid residues are coloured dark green. b, Detailed view of residues interacting with the bases of 3′3′-cGAMP. Parentheses indicate equivalent position in T4 Acb1. c, Conformations of 3′3′-cGAMP bound to STING (Protein Data Bank (PDB): 5CFM) or Acb1. d, Detailed view of catalytic residues. Parentheses indicate equivalent position in T4 Acb1. e, Thin-layer chromatography analysis of 3′3′-cGAMP cleavage by T4 Acb1 point mutants. Data are representative of three independent experiments. f, Schematic of reactions catalysed by Acb1 and Apyc1.