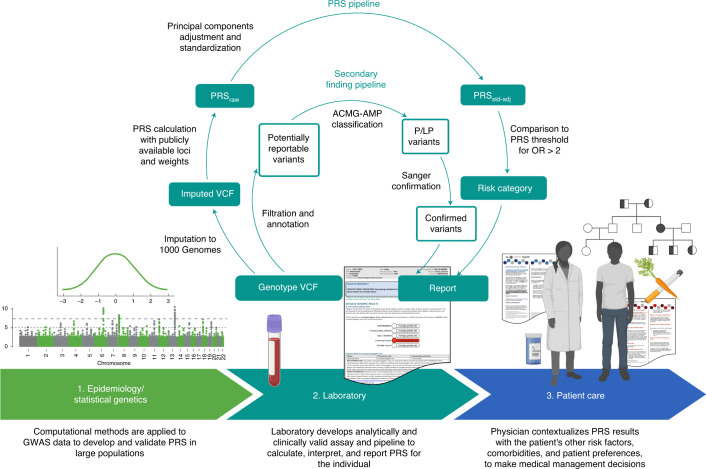
Fig. 1. Translation of PRS from discovery to the clinic, including a clinical PRS laboratory pipeline for prospectively collected samples.
In phase 1, PRS are developed, validated and compared to optimize performance in large populations. In phase 2, a clinical laboratory chooses publicly available PRS to implement and develop an analytically and clinically valid assay. For the GenoVA Study, genotype array data are imputed against 1000 Genomes Project data and used to calculate published PRS (PRSraw). PRSraw is adjusted for population structure and standardized as described in the text (PRSstd-adj). High-risk status for each disease is defined as PRS values above published thresholds for OR > 2. A parallel pipeline annotates and filters variants for potentially actionable pathogenic (P) and likely pathogenic (LP) variants in the ACMG SF v2.0 secondary finding gene list. Variants are manually classified according to American College of Medical Genetics and Genomics–Association for Molecular Pathology (ACMG-AMP) criteria by qualified laboratorians and confirmed using Sanger sequencing. Results from both components of the pipeline are included on the laboratory report. In phase 3 the treating physician uses the whole patient context to interpret the significance of the PRS for the patient’s health and healthcare management. Both the physician and patient will probably need educational and consultative support to make medical decisions based on PRS results.