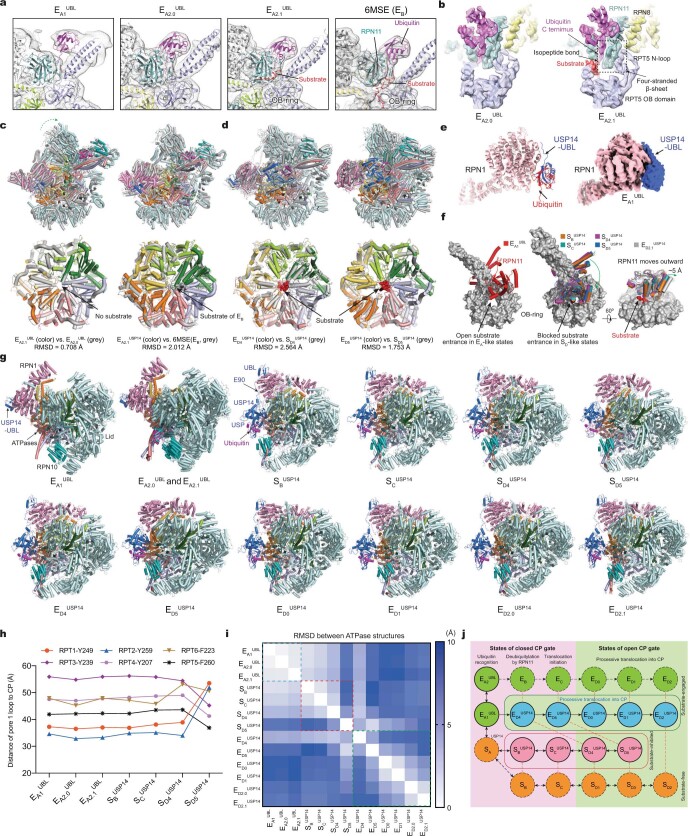
Extended Data Fig. 7. Structural comparison of the USP14-bound proteasome of different states.
a, Structural comparison of states , and shows the ubiquitin transfer from the RPT4-RPT5 coiled-coil (CC) domain to RPN11. The cryo-EM densities rendered as grey mesh representations are low-pass-filtered to 8 Å for visual clarity of comparison. b, Structural comparison of ubiquitin-RPN11-RPT5 interaction in state and . Cryo-EM densities rendered transparent surfaces are superimposed with the atomic models. The overall conformation of resides between and . Both states and exhibited RPN11-bound ubiquitin and no substrate densities in the AAA-ATPase motor. A short stretch of ubiquitin-linked substrate density is bound to the cleft between RPN11 and the OB ring in but not . In both states, the RPT5 N-loop (residues 99-119) pairs with one side of the Insert-1 β-hairpin of RPN11, the other side of which is paired with the C terminus of ubiquitin. They together form a four-stranded β-sheet, a feature that was previously visualized at atomic level only in state EB of the USP14-free proteasome10. c, Structural comparison of the RP and ATPase between and and between and EB (PDB ID 6MSE). d, Structural comparison of the RP and ATPase between and and between and . The root-mean-squared-deviation (RMSD) values for the ATPase components are shown below each panel of structural comparison in (c) and (d). e, Binding of UBL of USP14 to the T2 site of RPN1 in state as compared to the binding of ubiquitin to RPN1 in USP14-free state EA1 from previous studies10. Right inset shows the cryo-EM density of UBL-bound RPN1 in . f, Comparison of the RPN11-OB ring interface in different states. Left panel, the interface in the EA-like states shows an open OB ring for substrate entrance. Middle panel, the substrate entrance of the OB ring is blocked by RPN11 in the substrate-inhibited states. Right panel, RPN11 is rotated outward slightly by ~5 Å to make way for substrate translocation through the OB ring in state (grey cartoon) as compared to those in the substrate-inhibited states (colorful cartoon). g, Side-by-side structural comparison of the USP14-bound RP in all states from the top view showing differential rotation of the lid and RPN1. h, Plots of the distance of pore-1 loop to the CP for those states not shown in Fig. 3c. The comparison shows that the pore-loop staircase architecture in state , or is similar to that of EA-like state. i, Root-mean-squared-deviation (RMSD) values of the ATPase structures are mapped between any pairs of the thirteen states. j, An integrated schematic diagram of proteasome state transitions illustrates the full functional cycles of the proteasome in the presence and absence of USP14. The solid circles are the states observed in the current study, whereas the dashed circles are the states observed in previous studies of substrate-free8 (orange) or substrate-engaged human proteasome10 (green) in the absence of USP14. Color blue and salmon label the substrate-engaged and substrate-inhibited USP14-proteasome states, respectively. The states with closed and open CP gate are placed in pink and limon backgrounds, respectively. Vertical orange dashed lines link the state pairs with comparable AAA-ATPase structures. Black solid arrows and dashed arrows represent the possible structural transitions connecting the states observed in the current study and pervious USP14-free studies8,10, respectively