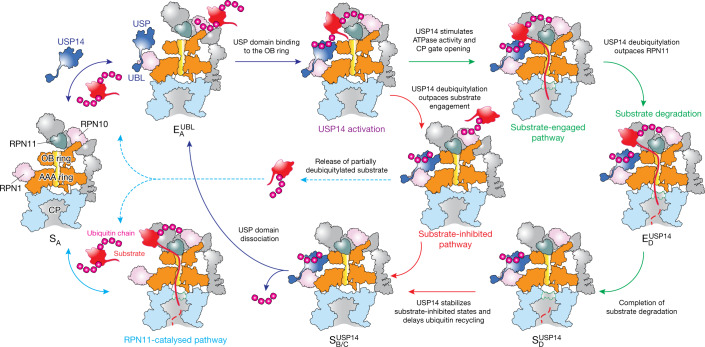
Fig. 4. Proposed model of USP14-mediated regulation of proteasome function.
USP14 binding to the RPN1 and RPT1 subunits of the proteasome primes USP14 activation, whereas ubiquitin–substrate conjugates recruited to the proteasome’s ubiquitin receptors facilitate ubiquitin recognition by USP14. RPN11-catalysed pathway (turquoise solid arrow) is allosterically excluded once USP14 is recruited to the proteasome (dark blue arrows). USP14 binding creates two parallel state-transition pathways of the proteasome. Along the substrate-inhibited pathway (red arrows), which has RPN11 blocking the substrate entrance of the OB ring before any substrate insertion takes place, USP14 trims ubiquitin chains and releases the substrate from the proteasome, thus preventing the substrate degradation (dashed turquoise arrows). Along the substrate-engaged pathway (green arrows), a substrate has already been inserted into the ATPase ring and RPN11 narrows down on the OB ring but does not block substrate translocation through the OB ring (Extended Data Fig. 7f). Although our data do not intuitively explain why USP14 trims ubiquitin until the last one on a substrate remains, the structures provide geometric constraints for polyubiquitin chain binding to both ubiquitin receptors and USP14 and suggest that ubiquitin recognition by USP14 in the proteasome requires at least one additional helper ubiquitin chain that is already anchored on a nearby ubiquitin receptor. This helper ubiquitin chain may not be available for USP14 binding but can be readily trimmed by RPN11.