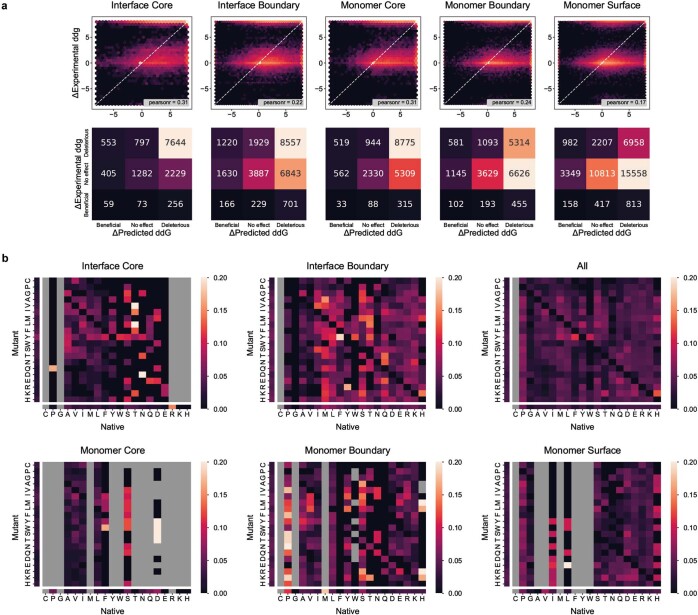
Extended Data Fig. 6. Computational analysis of the experimental SSM results.
a, Ability of Rosetta to predict mutational effects. This graph shows the observed experimental effect of each mutation versus Rosetta’s expected effect. For each plotted point, the delta refers to the effect versus the parent SSM design; therefore a “Beneficial” mutation is one that would improve affinity relative to the original designed protein the SSM was based on. The ΔExperimental ddg is derived from FACS data using the SC50 values (see Methods). Confidence intervals were collapsed to their center point to make this graph and “No effect” refers to mutations with less than a 1 kcal/mol change. Binder region definitions: Interface Core: residue contacts target protein and has no SASA (Solvent Accessible Surface Area) in bound state; Interface Boundary: residue contacts target protein, but does have SASA; Monomer Core: residue has no SASA and does not contact target; Monomer Boundary: residue has intermediate SASA and does not contact target; Monomer Surface: residue has full SASA and does not contact target. see Methods SSM Validation for further explanation. b, Mutations observed in SSM experiments that improved affinity bind at least 1kcal/mol graphed by relative frequency. Plotted is the #_times_Native_to_Mutant_improved_affinity / #_times_Native_to_Mutant_tested_in SSMs. A value of 0.10 with x-axis F and y-axis W could therefore represent that for 2 of 20 times W was substituted for Y, the affinity improved. Separated bars on each axis represent pooled data for the entire row/column. Grey boxes indicate mutations that occurred fewer than 5 times. Only SSM designs with a validation score of 0.005 or better were considered. While some cells are clipped, none extended beyond 0.25. Binder region definitions: Interface Core: residue contacts target protein and has no SASA in bound state; Interface Boundary: residue contacts target protein, but does have SASA; Monomer Core: residue has no SASA and does not contact target; Monomer Boundary: residue has intermediate SASA and does not contact target; Monomer Surface: residue has full SASA and does not contact target.