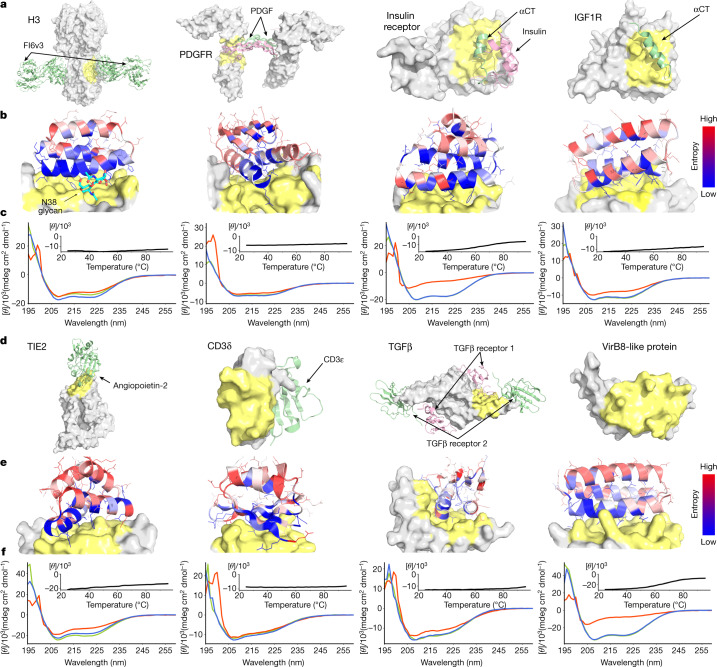
Fig. 2. De novo design and characterization of miniprotein binders.
a, d, Naturally occurring target protein structures shown in surface representation, with known interacting partners in cartoons where available. Regions targeted for binder design are coloured in pale yellow or green; the remainder of the target surface is in grey. See Extended Data Fig. 3 for side-by-side comparisons of the native binding partners and the computational design models. The PDB identifiers are 3ZTJ (H3), 3MJG (PDGFR), 4OGA (IR), 5U8R (IGF1R), 2GY7 (TIE2), 1XIW (CD3δ), 3KFD (TGFβ) and 4O3V (VirB8). αCT, α-chain C-terminal helix. b, e, Computational models of designed complexes coloured by site saturation mutagenesis results. Designed binding proteins (cartoons) are coloured by positional Shannon entropy, with blue indicating positions of low entropy (conserved) and red those of high entropy (not conserved); the target surface is in grey and yellow. The core residues and binding interface residues are more conserved than the non-interface surface positions, consistent with the computational models. Full SSM maps over all positions of all the de novo designs are provided in the Supplementary Information. c, f, Circular dichroism spectra at different temperatures (green, 25 °C; red, 95 °C; blue, 95 °C followed by 25 °C), and circular dichroism signals at 222-nm wavelength as a function of temperature for the optimized designs (insets). See Extended Data Fig. 4 for the biolayer interferometry characterization results of the optimized designs.