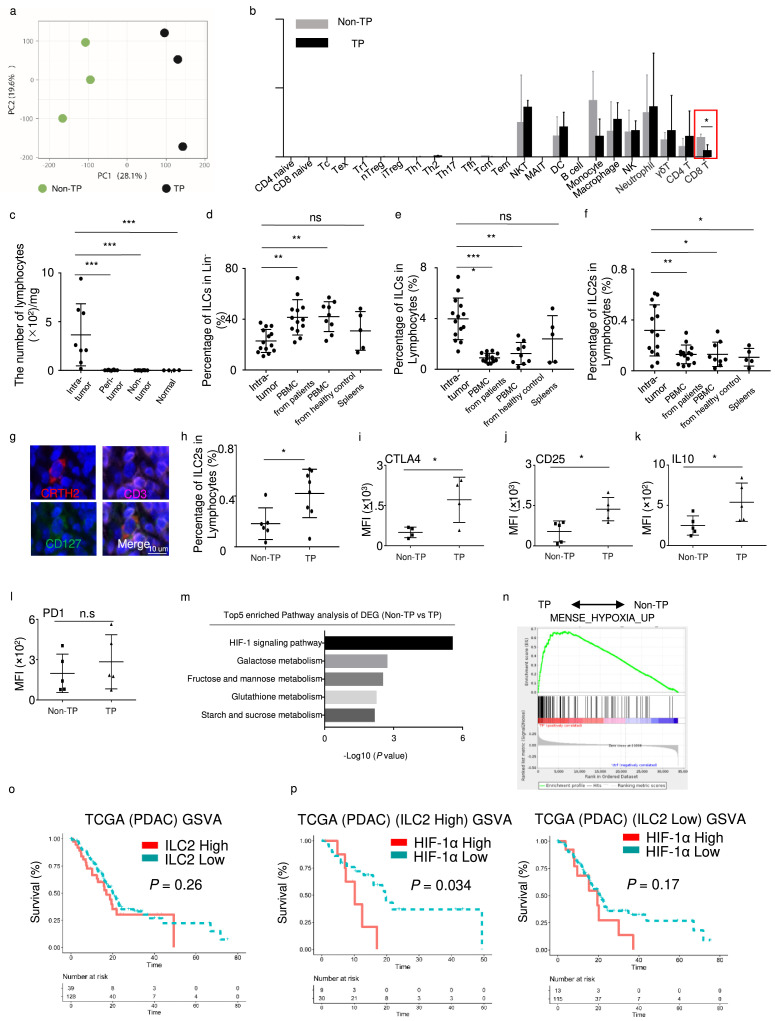
Figure 1.
Characterization of a novel ILC2 group in pancreatic cancer.
(a) Visualization of TP (n = 3) and Non-TP (n = 3) in PCA, computed on a gene expression matrix. (b) Major immune cells abundance in TP and Non-TP patients estimated by ImmuCellAI algorithm, using microarray data. (c) Lymphocyte numbers in intra-tumor (n = 8), peri-tumor (n = 8), non-tumor (n = 8), and normal tissues (n = 4), assessed with the Mann-Whitney U test. (d) Percentages of ILCs in Lin− in tumors (n = 14), PBMCs from patients (n = 13), PBMCs from healthy controls (n = 9), and spleens (n = 5). (e) Percentages of ILCs in lymphocytes from tumors (n = 14), PBMCs from patients (n = 13), PBMCs from healthy controls (n = 9), and spleens (n = 5). (f) Percentages of ILC2s in lymphocytes from tumors (n = 14), PBMCs from patients (n = 13), PBMCs from healthy controls (n = 9), and spleens (n = 5). (g) Representative immunofluorescence images (magnification, × 1500) of human PDAC tissue stained with antibodies against CRTH2 (red), CD3 (pink), and CD127 (green). Nuclei were stained with DAPI (blue). (h) Percentages of ILC2s in lymphocytes from Non-TP (n = 6) and TP (n = 8) patients. Statistical analysis of CTLA4 (n = 4) (i), CD25 (n = 5) (j), IL10 (n = 5) (k) and PD1 (n = 5) (l) in ILC2 from Non-TP and TP patients. (m) Pathway analysis of differentially expressed genes highly expressed in TP. (n) Gene set enrichment analysis (GSEA) of hypoxia pathway in TP and Non-TP patients. (o) Gene Set Variance Analysis (GSVA) using a 12-gene signature of ILC2s derived from single-cell sequencing analysis. Optimal cutoff point was determined by the survminer R package. Analysis of survival prediction within PDAC TCGA cohort. (p) Survival prediction of PDAC according to HIF-1α expression in “ILC2 high” or “ILC2 low” patients’ group. Data are expressed as means ± SD and compared using the unpaired t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).