Figure 4:
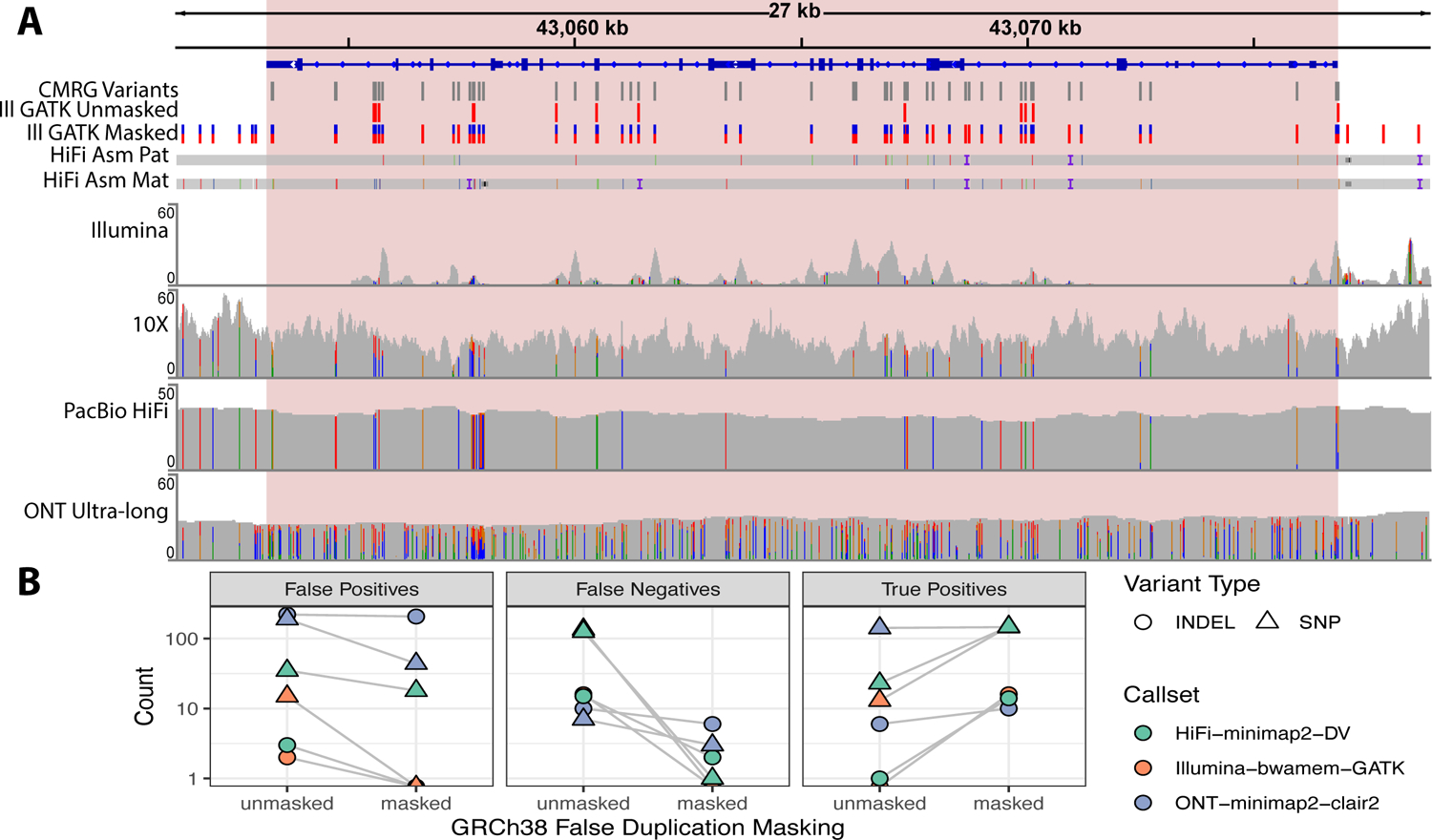
(A) The benchmark resolves the gene CBS, which has a highly homologous gene CBSL due to a false duplication in GRCh38 that is not in HG002 or GRCh37. The duplication in GRCh38 causes Illumina and PacBio HiFi reads from one haplotype to mismap to CBSL instead of CBS. The ultralong ONT reads, 10x Genomics linked reads, and assembled PacBio HiFi contigs map properly to this region for both haplotypes because they contain sufficient flanking sequence. When the falsely duplicated sequence is masked using our new version of GRCh38, variant calls from a standard Illumina-GATK pipeline (ILMN-GATK w/ Mask VCF) are completely concordant with the new benchmark. Pink shaded box indicates CMRG benchmark regions, only variants within the benchmark regions are included in the benchmark. (B) Comparison of variant accuracy for GRCh38 before and after masking false duplications on chromosome 21. The new benchmark demonstrates decreases in false negative and false positive errors for 3 callsets in the falsely duplicated genes CBS, CRYAA, and KCNE1 when mapping to the masked GRCh38.
