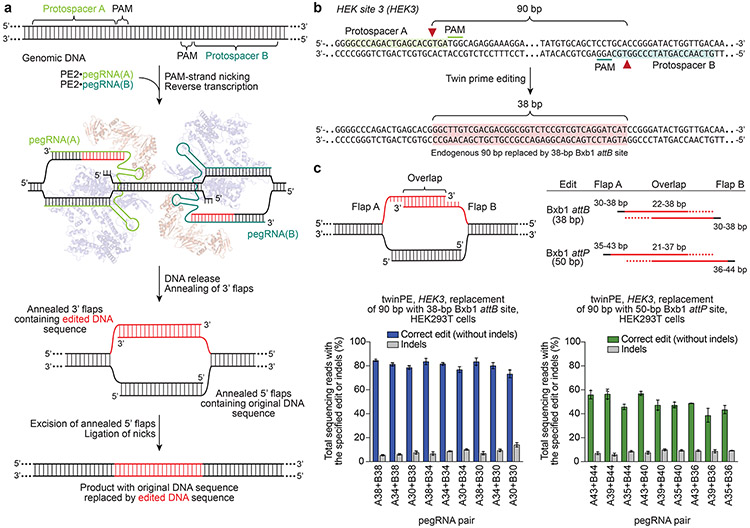
Figure 1 ∣. Overview of twinPE and twinPE-mediated sequence replacement.
(a) TwinPE systems target genomic DNA sequences that contain two protospacer sequences on opposite strands of DNA. PE2•pegRNA complexes target each protospacer, generate a single-stranded nick, and reverse transcribe the pegRNA-encoded template containing the desired insertion sequence. After synthesis and release of the 3’ DNA flaps, a hypothetical intermediate exists possessing annealed 3’ flaps containing the edited DNA sequence and annealed 5’ flaps containing the original DNA sequence. Excision of the original DNA sequence contained in the 5’ flaps, followed by ligation of the 3’ flaps to the corresponding excision sites, generates the desired edited product. (b) Example of twinPE-mediated replacement of a 90-bp sequence in HEK3 with a 38-bp Bxb1 attB sequence. Red arrows indicate the position of pegRNA-induced nicks. (c) Evaluation of twinPE in HEK293T cells for the installation of the 38-bp Bxb1 attB site as shown in (b) or the 50-bp Bxb1 attP site at HEK3 using pegRNAs that template varying lengths of the insertion sequence. pegRNA names indicate spacer (A or B) and length of RT template. Values and error bars reflect the mean and s.d. of three independent biological replicates.