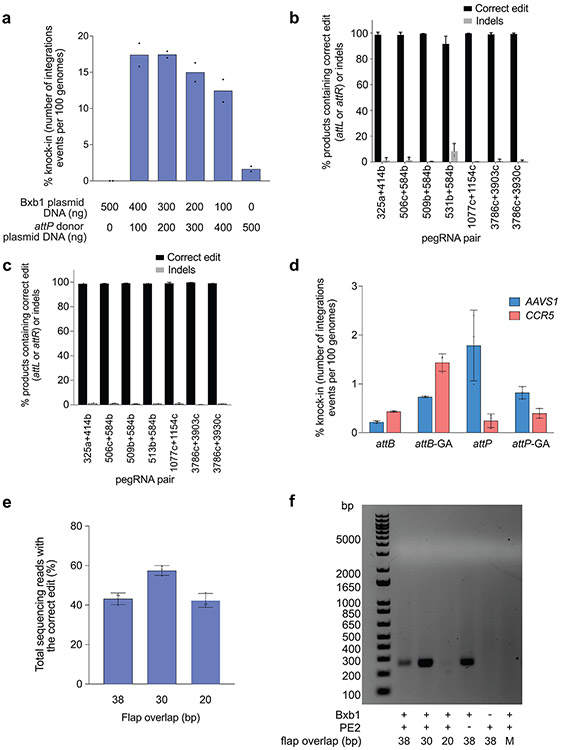
Extended Data Fig. 6. TwinPE combined with Bxb1 recombinase for targeted knock-in of donor DNA plasmids.
(a) Bxb1-mediated DNA donor knock-in in clonal HEK293T cell lines. Transfection of a HEK293T clonal cell line containing homozygous attB site insertion at CCR5 with varying amounts of Bxb1-expressing plasmid and attP-containing donor DNA plasmid. Knock-in efficiency was quantified by ddPCR. Values and error bars reflect the mean of two independent biological replicates. (b) Assessment of genome-donor junction purity by high-throughput sequencing. Genomic DNA from single-transfection knock-in experiments was amplified with a forward primer that binds the genome and a reverse primer that binds within the donor plasmid (Supplementary Table 2). Values and error bars reflect the mean and s.d. of three independent biological replicates. (c) Assessment of genome-donor junction purity at the other junction by high-throughput sequencing as performed in (b). Values and error bars in 506c+584b,509b+584b, 1077c+1154c, and 3786c+3903c reflect the mean and s.d. of three independent biological replicates. Values in 325a+414b, 513b+584b, 3786c+3930c reflect the mean of two independent biological replicates. (d) Multiplexed single-transfection knock-in at AAVS1 and CCR5. HEK293T cells were transfected with plasmids encoding PE2, Bxb1, a pair of pegRNAs for the insertion of attP at AAVS1, an attB-donor, a pegRNA pair for the insertion of one of four attachment sites (attB, attB-GA, attP, or attP-GA) at CCR5, and a corresponding donor. Knock-in was observed at both target loci under all four conditions. Insertion of attP at AAVS1 and attB at CCR5 gave the lowest knock-in efficiencies overall (0.2% at AAVS1, 0.4% at CCR5). Insertion of attP at both sites yielded the highest levels of knock-in at AAVS1 (1.8%) but low levels (0.2%) at CCR5. When an orthogonal edit (attB-GA or attP-GA) was introduced at CCR5, AAVS1 knock-in was 0.7-0.8%. Higher knock-in at CCR5 was observed with attB-GA (1.4%) than with attP-GA (0.4%), consistent with our single locus knock-in results. Values and error bars reflect the mean and s.d. of three independent biological replicates. (e) and (f) Effects of reducing pegRNA overlap on twinPE efficiency and donor/pegRNA recombination. (e) The editing efficiencies of pairs of pegRNAs for insertion of Bxb1 attB at CCR5 were measured by high-throughput sequencing. The pairs differed in the amount of overlap shared between their flaps, from 38 bp (full-length attB sequence) down to 20 bp. Editing efficiency of the pairs with shorter overlaps was comparable to the pair with full-length overlap. Values and error bars reflect the mean and s.d. of three independent biological replicates. (f) Assessment of recombination between attB-containing pegRNA plasmids and attP-containing donor plasmids. Following transfection of HEK293T cells with the indicated samples, isolated DNA was amplified with a forward primer that binds the pegRNA expression plasmid (TTGAAAAAGTGGCACCGAGT) and a reverse primer that binds the donor plasmid (CTCCCACTCATGATCTA). A positive 256-bp PCR band confirms recombination between the two plasmids. When the pegRNA encodes full-length attB (38-bp) or a truncated version of attB with 30-bp of overlap between flaps, a band is observed; however, recombination is not observed when the pegRNAs encode a truncated attB with only 20-bp of flap overlap. The “No PE2” control uses the 38-bp overlap pegRNA pair. No recombination is observed in the absence of Bxb1 or if the donor and pegRNA plasmids both bear attB (Mismatch, “M”). Three independent biological replicates were performed and a representative image from one of the replicates is shown.