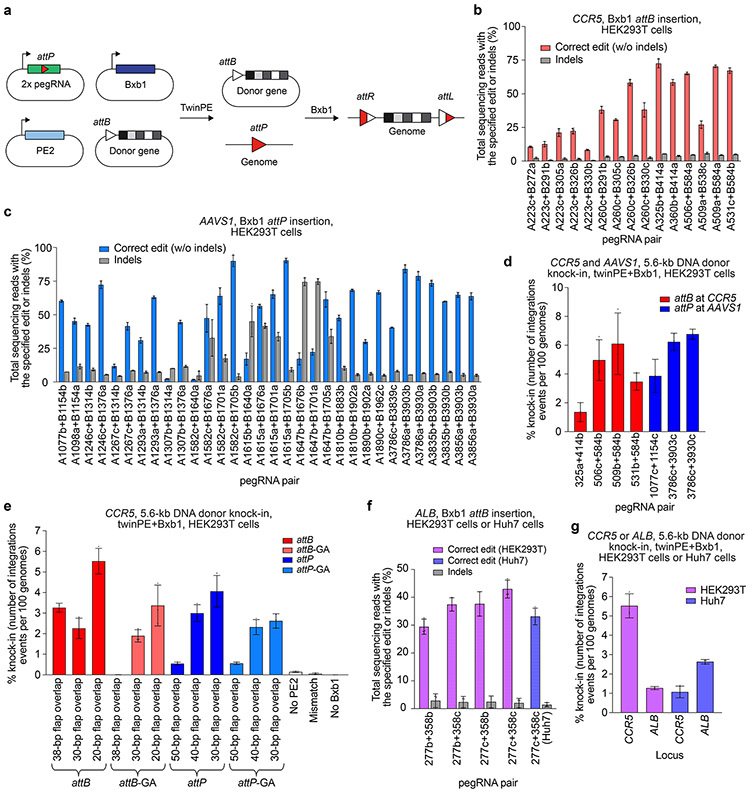
Figure 3 ∣. Site-specific genomic integration of DNA cargo with twinPE and Bxb1 recombinase in human cells.
(a) Schematic diagram of twinPE and Bxb1 recombinase-mediated site-specific genomic integration of DNA cargo. (b) Screening of twinPE pegRNA pairs for insertion of the Bxb1 attB sequence at the CCR5 locus in HEK293T cells. (c) Screening of twinPE pegRNA pairs for installation of the Bxb1 attP sequence at the AAVS1 locus in HEK293T cells. (d) Single transfection knock-in of 5.6-kb DNA donors using twinPE pegRNA pairs targeting CCR5 (red) or AAVS1 (blue). The twinPE pegRNAs install attB at CCR5 or attP at AAVS1. Bxb1 integrates a donor bearing the corresponding attachment site into the genomic attachment site. The number of integration events per 100 genomes is defined as the ratio of the target amplicon spanning the donor-genome junction to a reference amplicon in ACTB, as determined by ddPCR. (e) Optimization of single-transfection integration at CCR5 using the A531+B584 spacers for the twinPE pegRNA pair. Identity of the templated edit (attB or attP), identity of the central dinucleotide (wild-type GT or orthogonal mutant GA), and length of the overlap between flaps were varied to identify combinations that supported the highest integration efficiency. % knock-in quantified as in (d). (f) Pairs of pegRNAs were assessed for their ability to insert Bxb1 attB into the first intron of ALB. Protospacer sequences (277 and 358) are constant across the pegRNA pairs. The pegRNAs vary in their PBS lengths (variant b or c). The 277c/358c pair that performs best in HEK293T cells can also introduce the desired edit in Huh7 cells. (g) Comparison of single transfection knock-in efficiencies at CCR5 and ALB in HEK293T and Huh7 cell lines. % knock-in quantified as in (d). Values and error bars reflect the mean and s.d. of three independent biological replicates.