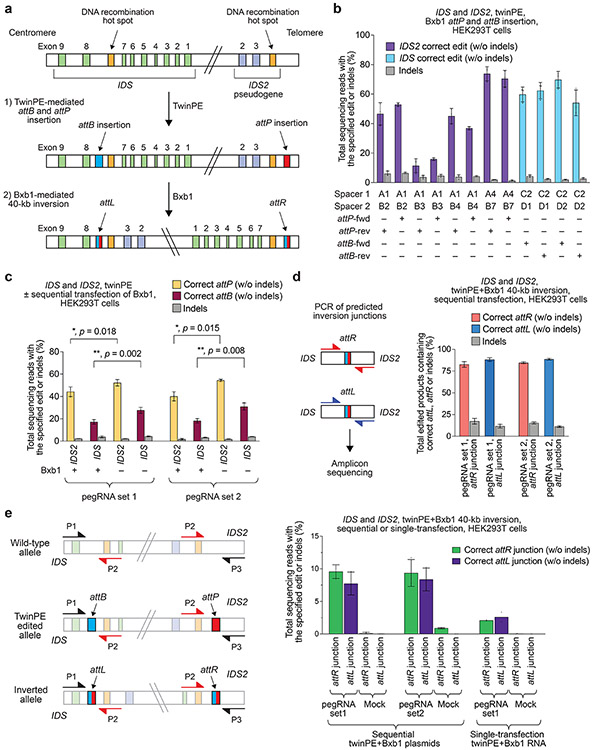
Figure 4 ∣. Site-specific large genomic sequence inversion with twinPE and Bxb1 recombinase in human cells.
(a) Schematic diagram of DNA recombination hot spots in IDS and IDS2 that lead to pathogenic 39-kb inversions, and the combined twinPE-Bxb1 strategy for installing or correcting the IDS inversion. (b) Screen of pegRNA pairs at IDS and IDS2 for insertion of attP or attB recombination sites. Values and error bars reflect the mean and s.d. three independent biological replicates. (c) DNA sequencing analysis of the IDS and IDS2 loci after twinPE-mediated insertion of attP or attB sequences, with or without subsequent transfection with Bxb1 recombinase. P-values were derived from a Student’s two-tailed t-test. Values and error bars reflect the mean and s.d. three independent biological replicates. (d) 40,167-bp IDS inversion product purities at the anticipated inversion junctions after twinPE-mediated attachment site installation and sequential transfection with Bxb1 recombinase. Values and error bars reflect the mean and s.d. three independent biological replicates. (e) Analysis of inversion efficiency by amplicon sequencing at IDS and IDS2 loci after sequential transfection or single-step transfection of twinPE editing components and Bxb1 recombinase. Values and error bars for sequential transfection reflect the mean and s.d. of three independent biological replicates; values for single-transfection reflect the mean of two independent biological replicates.