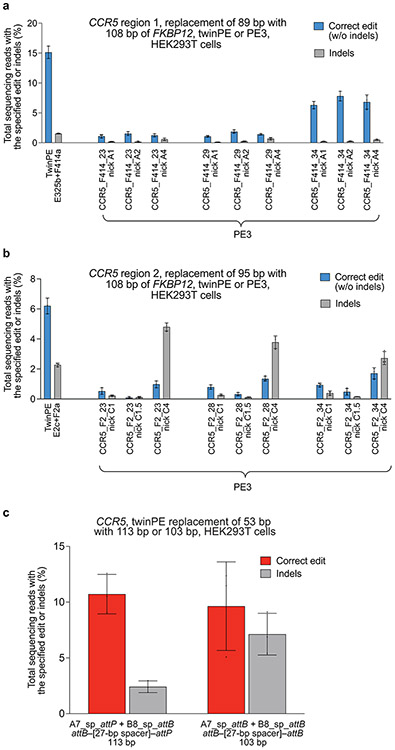
Extended Data Fig. 1. Twin prime editing mediates sequence replacements at CCR5.
(a) Replacement of endogenous sequence within CCR5 region 1 with a 108-bp fragment of FKBP12 cDNA using twinPE (FKBP12 sequence oriented in the forward direction,) or PE3 (FKBP12 sequence oriented in the reverse direction). For PE3 editing, pegRNA RT templates were designed to encode 108 base pairs of FKBP12 cDNA sequence and one of three different target-site homology sequence lengths. For PE3 edits, each pegRNA was tested with three nicking sgRNAs. (b) Replacement of endogenous sequence within CCR5 region 2 with a 108-bp fragment of FKBP12 cDNA sequence using twinPE (FKBP12 sequence oriented in the forward direction) or PE3 (FKBP12 sequence oriented in the reverse direction). As in (a), PE3 edits were tested with pegRNAs containing RT templates that were designed to encode 108 base pairs of FKBP12 cDNA sequence and one of three different target-site homology sequence lengths. For PE3 edits, each pegRNA was tested with three nicking sgRNAs. Values and error bars reflect the mean and s.d. of three independent biological replicates. (c) Transfection of HEK293T cells with a pair of pegRNAs targeting CCR5 leads to replacement of 53 base pairs of endogenous sequence with 113 base pairs (attB–[27-bp spacer]–attP) or 103 base pairs (attB–[27-bp spacer]–attB) of exogenous sequence. Values and error bars reflect the mean and s.d. of three independent biological replicates.