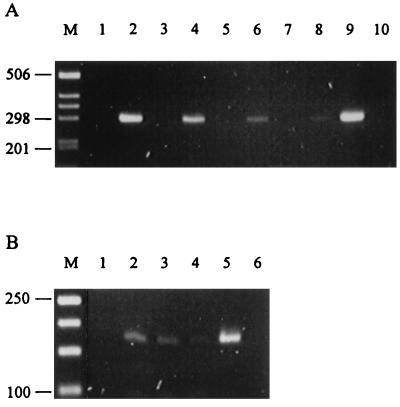
FIG. 3.
Amplification of Epichloë microsatellite loci from endophyte-infected plant material. Ethidium bromide-stained agarose (3% NuSieve) gels of PCR products amplified from genomic DNA isolated from grass tissue by using primers to microsatellite loci B1 (A) and B10 (B). (A) PCR products generated with DNA prepared from AR1-infected L. perenne × L. multiflorum hybrid ryegrass (lanes 2, 4, 6, and 8) and endophyte-free hybrid ryegrass (lanes 1, 3, 5, and 7) by the CTAB method (18) at total DNA concentrations of 2 ng μl−1 (lanes 1 and 2), 400 pg μl−1 (lanes 3 and 4), 200 pg μl−1 (lanes 5 and 6), and 40 pg μl−1 (lanes 7 and 8); at an AR1 fungal genomic DNA concentration of 40 pg μl−1 (lane 9); and with no DNA (lane 10). Lane M, 1-kb ladder. (B) PCR products generated with DNA prepared from AR1-infected L. perenne × L. multiflorum hybrid ryegrass by the FastDNA Kit H minipreparation method at total DNA concentrations of 2 ng μl−1 (lane 1), 400 pg μl−1 (lane 2), 80 pg μl−1 (lane 3), and 16 pg μl−1 (lane 4); at an AR1 fungal genomic DNA concentration of 40 pg μl−1 (lane 5); and with no DNA (lane 6). Lane M, 50-bp ladder. The numbers on the left of the figures indicate the sizes in base pairs of the DNA size markers.