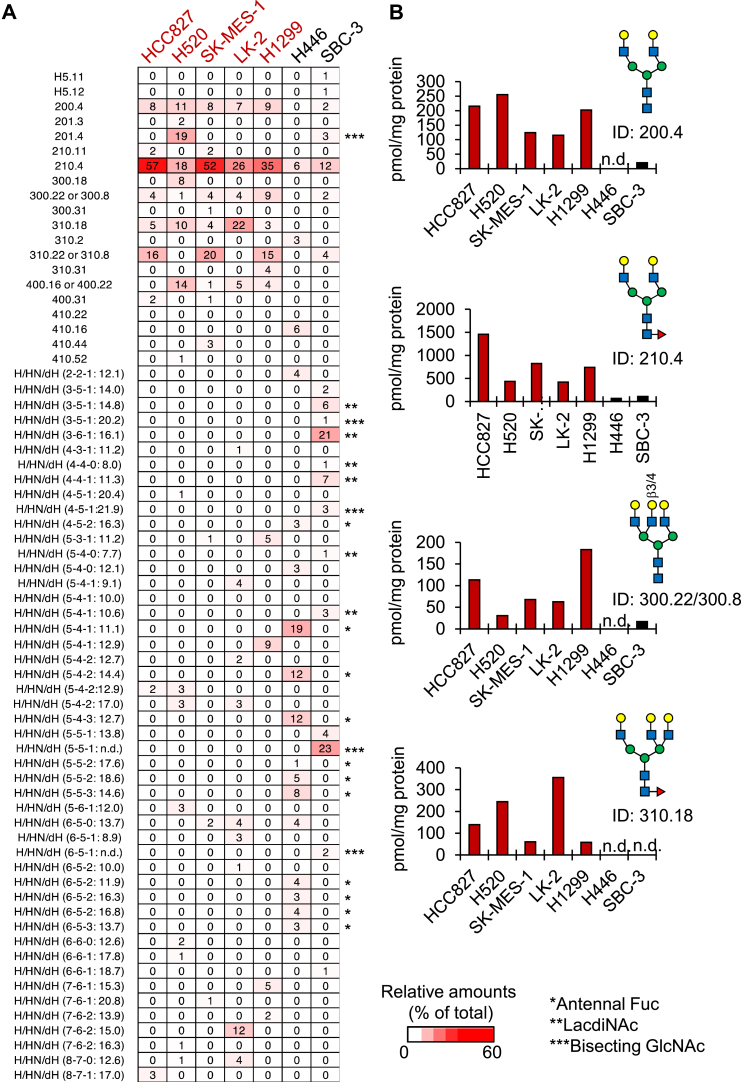
Figure 2.
SCLC-sEVs and NSCLC-sEVs have different N-glycan profiles.A, the relative amounts of N-glycans in NSCLC-sEVs (H520, SK-MES-1, LK-2, H1299, and HCC827) and SCLC-sEVs (H446 and SBC-3) were shown as a heatmap. Glycan IDs for deduced N-glycan structures are based on the GALAXY database. Glycan IDs for undeduced N-glycan structures are indicated as the number of H/HN/dH (x-y-z), followed by their GU in reversed-phase HPLC. n.d., not determined. Asterisks indicate N-glycans with antennal fucose (∗), LacdiNAc (∗∗), and bisecting GlcNAc (∗∗∗). B, N-glycans that were enriched in NSCLC-sEVs were extracted from a heatmap in (A), and the amounts were expressed as pmol/mg protein of sEVs. n.d., not detected. See also Figs S2–S5 and Data S1. Three batches of sEVs were independently prepared, fluorescently labeled, and analyzed by ODS-HPLC after desialylation, and similar HPLC profiles were observed among three batches. Rigorous structural analysis by MS was performed once for the representative sample. LacdiNAc, N-acetylgalactosamine-GlcNAc; NSCLC, non–small-cell lung carcinoma; SCLC, small-cell lung carcinoma; sEV, small extracellular vesicle.