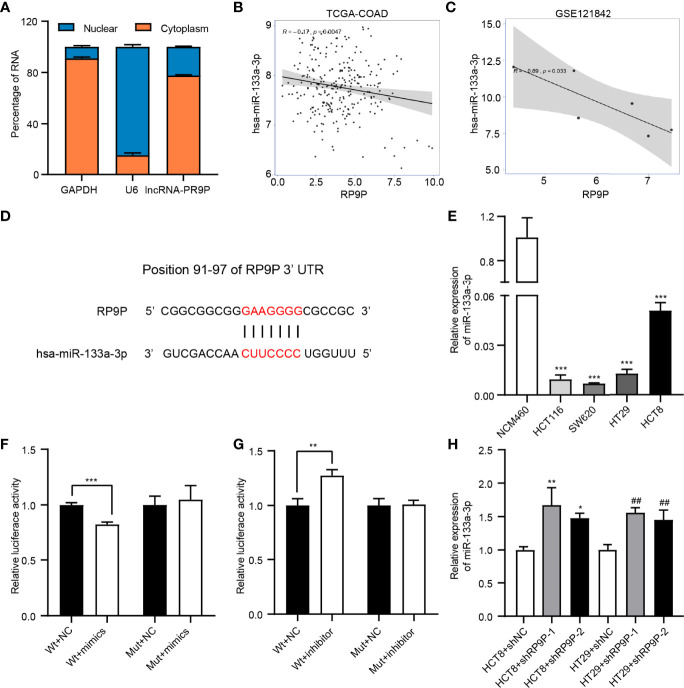
Figure 4.
Identification of miR-133a-3p as a direct target gene of RP9P. (A) Subcellular localization of RP9P in colorectal cancer (CRC) cells. (B, C) The relationship between RP9P and miR-133a-3p was determined using TCGA-COADREAD (B) and GSE121842 (C) data. (D) miR-133a-3p level in CRC cells identified using real-time (RT)-PCR (***p < 0.001 vs. NCM460 cells). (E) Predicted binding sites of RP9P and miR-133a-3p and mutated sites in the 3′-untranslated region (UTR) of RP9P. (F, G) Luciferase activity in nonsense control (NC)-, miR-133a-3p mimic- (F), and miR-133a-3p inhibitor (G)‐treated CRC cells with wild-type or mutant RP9P 3′-UTR gene fragments (**p < 0.01, ***p < 0.001). (H) The miR-133a-3p level in CRC cells with RP9P silenced was examined using RT-PCR (*p < 0.05, **p < 0.01 vs. HCT8 + shNC; ## p < 0.01 vs. HT29 + shNC).