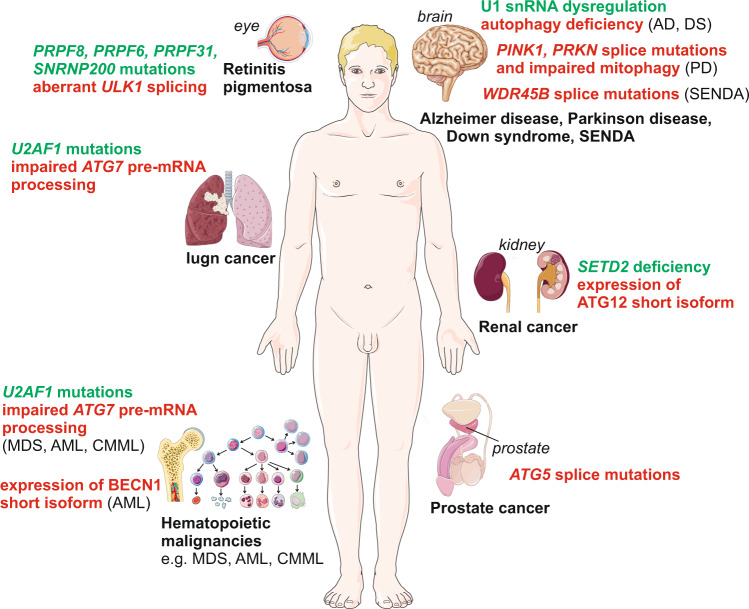
Fig. 4. Human diseases associated with alternative-splicing dependent autophagy impairment.
For details, see text body. In brief, U1 snRNP enrichment in familial Alzheimer disease (AD) and Down syndrome (DS) are associated with impaired autophagy. De novo WDR45/WIPI4 splice mutations and lowered autophagic flux are associated with static encephalopathy of childhood with neurodegeneration in adulthood (SENDA). Splicing mutations in PINK1 and PRKN/PARKIN lead to mitophagy impairment and contribute to PD and various cancer. Aberrant alternative splicing of ULK1 and impaired mitophagy associated with PRPF8, PRPF6, PRPF31 and SNRNP200 mutations contribute to retinitis-pigmentosa. U2AF1-mutation dependent ATG7 expression reduction, due to impaired pre-mRNA processing, in hematopoietic cells promotes autophagy impairment and oncogenic transformation, as observed in myelodysplastic syndrome (MDS), acute myeloid leukemia (AML), and chronic myelomonocytic leukemia (CMML) as well as lung adenocarcinoma. BECN1 short isoform generated by alternative splicing acts as a negative modulator of autophagy in AML. Deficiency in SETD2, an epigenetic regulator of alternative splicing, promotes the expression of an ATG12 short isoform and alteration of autophagic flux in renal cancer. Splice-site mutations in ATG5 lead to a dysregulation of autophagy in prostate cancer. Images from Servier Medical Art (smart.servier.com) licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/) were used to generate the depicted original illustration (smart.servier.com/terms-of-use/).