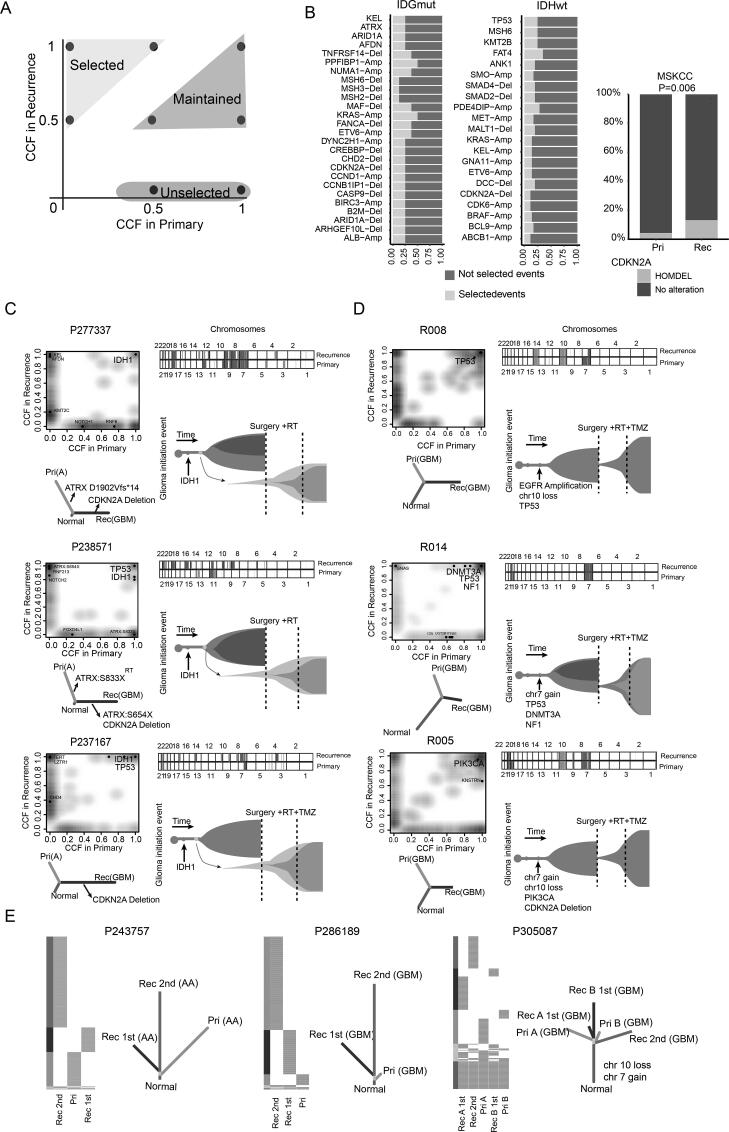
Fig. 3.
Evolution patterns of recurrence in gliomas. (A) Schematic illustration for the definition of selected, maintained, or unselected alterations in recurrences. (B) The stacked bar plots showing the proportions of recurrence-selected alterations that occurred in more than two primary-recurrence pairs. Proportions of CDKN2A deletion between primary and recurrent IDH-mut gliomas in MSKCC. (C-D) Genetic divergence between primary and recurrent tumors. Two-dimensional density plot based on CCFs of all SNVs identified at the primary tumor (x-axis) and recurrence (y-axis). The dot represented the driver genes. Chromosomal copy-number plots for paired glioma samples; in each plot, light red = gain; red = Amplification; light blue = loss; blue = Deletion. Phylogenetic trees were generated by maximum parsimony based on mutational presence or absence, for which canonical glioma drivers genes are labeled. (E) Phylogenetic trees depicted the evolution patterns in samples with multiple recurrences. RT: Radiotherapy; TMZ: Temozolomide; Pri: Primary gliomas; Rec: Recurrent gliomas. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)